Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation
- PMID: 23892457
- PMCID: PMC3746202
- DOI: 10.1038/emboj.2013.167
Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation
Abstract
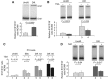
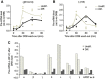
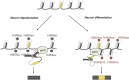
Alternative splicing contributes to cell type-specific transcriptomes. Here, we show that changes in intragenic chromatin marks affect NCAM (neural cell adhesion molecule) exon 18 (E18) alternative splicing during neuronal differentiation. An increase in the repressive marks H3K9me2 and H3K27me3 along the gene body correlated with inhibition of polymerase II elongation in the E18 region, but without significantly affecting total mRNA levels. Treatment with the general DNA methylation inhibitor 5-azacytidine and BIX 01294, a specific inhibitor of H3K9 dimethylation, inhibited the differentiation-induced E18 inclusion, pointing to a role for repressive marks in sustaining NCAM splicing patterns typical of mature neurons. We demonstrate that intragenic deployment of repressive chromatin marks, induced by intronic small interfering RNAs targeting NCAM intron 18, promotes E18 inclusion in undifferentiated N2a cells, confirming the chromatin changes observed upon differentiation to be sufficient to induce alternative splicing. Combined with previous evidence that neuronal depolarization causes H3K9 acetylation and subsequent E18 skipping, our results show how two alternative epigenetic marks regulate NCAM alternative splicing and E18 levels in different cellular contexts.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

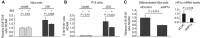

Similar articles
-
Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing.Proc Natl Acad Sci U S A. 2009 Mar 17;106(11):4325-30. doi: 10.1073/pnas.0810666106. Epub 2009 Feb 26. Proc Natl Acad Sci U S A. 2009. PMID: 19251664 Free PMC article.
-
Chromatin and alternative splicing.Cold Spring Harb Symp Quant Biol. 2010;75:103-11. doi: 10.1101/sqb.2010.75.023. Epub 2011 Feb 2. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21289049
-
Alternative Splicing of G9a Regulates Neuronal Differentiation.Cell Rep. 2016 Mar 29;14(12):2797-808. doi: 10.1016/j.celrep.2016.02.063. Epub 2016 Mar 17. Cell Rep. 2016. PMID: 26997278
-
Chromatin's thread to alternative splicing regulation.Chromosoma. 2013 Dec;122(6):465-74. doi: 10.1007/s00412-013-0425-x. Epub 2013 Aug 3. Chromosoma. 2013. PMID: 23912688 Review.
-
Chromatin and epigenetic regulation of pre-mRNA processing.Hum Mol Genet. 2012 Oct 15;21(R1):R90-6. doi: 10.1093/hmg/dds353. Epub 2012 Aug 29. Hum Mol Genet. 2012. PMID: 22936691 Free PMC article. Review.
Cited by
-
Epigenetic control of skeletal muscle atrophy.Cell Mol Biol Lett. 2024 Jul 8;29(1):99. doi: 10.1186/s11658-024-00618-1. Cell Mol Biol Lett. 2024. PMID: 38978023 Free PMC article. Review.
-
Splicing and transcription touch base: co-transcriptional spliceosome assembly and function.Nat Rev Mol Cell Biol. 2017 Oct;18(10):637-650. doi: 10.1038/nrm.2017.63. Epub 2017 Aug 9. Nat Rev Mol Cell Biol. 2017. PMID: 28792005 Free PMC article. Review.
-
H3K4 demethylase KDM5B regulates global dynamics of transcription elongation and alternative splicing in embryonic stem cells.Nucleic Acids Res. 2017 Jun 20;45(11):6427-6441. doi: 10.1093/nar/gkx251. Nucleic Acids Res. 2017. PMID: 28402433 Free PMC article.
-
Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms.Nucleic Acids Res. 2014 Jan;42(2):701-13. doi: 10.1093/nar/gkt875. Epub 2013 Sep 29. Nucleic Acids Res. 2014. PMID: 24081581 Free PMC article. Review.
-
Downregulation of SNRPG induces cell cycle arrest and sensitizes human glioblastoma cells to temozolomide by targeting Myc through a p53-dependent signaling pathway.Cancer Biol Med. 2020 Feb 15;17(1):112-131. doi: 10.20892/j.issn.2095-3941.2019.0164. Cancer Biol Med. 2020. PMID: 32296580 Free PMC article.
References
-
- Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR (2009) Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 - PubMed
-
- Alló M, Schor IE, Munoz MJ, de la Mata M, Agirre E, Valcarcel J, Eyras E, Kornblihtt AR (2010) Chromatin and alternative splicing. Cold Spring Harb Symp Quant Biol 75: 103–111 - PubMed
-
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsché E, Harel-Bellan A (2012) Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

