A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal
- PMID: 23892456
- PMCID: PMC3746198
- DOI: 10.1038/emboj.2013.161
A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal
Abstract
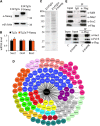
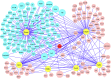
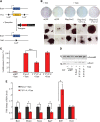
Embryonic stem (ES) cell self-renewal efficiency is determined by the Nanog protein level. However, the protein partners of Nanog that function to direct self-renewal are unclear. Here, we identify a Nanog interactome of over 130 proteins including transcription factors, chromatin modifying complexes, phosphorylation and ubiquitination enzymes, basal transcriptional machinery members, and RNA processing factors. Sox2 was identified as a robust interacting partner of Nanog. The purified Nanog-Sox2 complex identified a DNA recognition sequence present in multiple overlapping Nanog/Sox2 ChIP-Seq data sets. The Nanog tryptophan repeat region is necessary and sufficient for interaction with Sox2, with tryptophan residues required. In Sox2, tyrosine to alanine mutations within a triple-repeat motif (S X T/S Y) abrogates the Nanog-Sox2 interaction, alters expression of genes associated with the Nanog-Sox2 cognate sequence, and reduces the ability of Sox2 to rescue ES cell differentiation induced by endogenous Sox2 deletion. Substitution of the tyrosines with phenylalanine rescues both the Sox2-Nanog interaction and efficient self-renewal. These results suggest that aromatic stacking of Nanog tryptophans and Sox2 tyrosines mediates an interaction central to ES cell self-renewal.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
Distinct Contributions of Tryptophan Residues within the Dimerization Domain to Nanog Function.J Mol Biol. 2017 May 19;429(10):1544-1553. doi: 10.1016/j.jmb.2016.12.001. Epub 2016 Dec 6. J Mol Biol. 2017. PMID: 27939294 Free PMC article.
-
Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma.J Biol Chem. 2012 Sep 21;287(39):32800-24. doi: 10.1074/jbc.M111.308528. Epub 2012 Jul 30. J Biol Chem. 2012. PMID: 22847005 Free PMC article.
-
SUMOylation represses Nanog expression via modulating transcription factors Oct4 and Sox2.PLoS One. 2012;7(6):e39606. doi: 10.1371/journal.pone.0039606. Epub 2012 Jun 22. PLoS One. 2012. PMID: 22745796 Free PMC article.
-
Pluripotency transcription factors and cancer stem cells: small genes make a big difference.Chin J Cancer. 2013 Sep;32(9):483-7. doi: 10.5732/cjc.012.10282. Epub 2013 Feb 19. Chin J Cancer. 2013. PMID: 23419197 Free PMC article. Review.
-
Nanog and transcriptional networks in embryonic stem cell pluripotency.Cell Res. 2007 Jan;17(1):42-9. doi: 10.1038/sj.cr.7310125. Cell Res. 2007. PMID: 17211451 Review.
Cited by
-
PHC1 maintains pluripotency by organizing genome-wide chromatin interactions of the Nanog locus.Nat Commun. 2021 May 14;12(1):2829. doi: 10.1038/s41467-021-22871-0. Nat Commun. 2021. PMID: 33990559 Free PMC article.
-
Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells.Nature. 2018 Apr;556(7702):510-514. doi: 10.1038/s41586-018-0048-8. Epub 2018 Apr 18. Nature. 2018. PMID: 29670286 Free PMC article.
-
Mapping the route from naive pluripotency to lineage specification.Philos Trans R Soc Lond B Biol Sci. 2014 Dec 5;369(1657):20130540. doi: 10.1098/rstb.2013.0540. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25349449 Free PMC article. Review.
-
Pluripotency Factors on Their Lineage Move.Stem Cells Int. 2016;2016:6838253. doi: 10.1155/2016/6838253. Epub 2015 Dec 6. Stem Cells Int. 2016. PMID: 26770212 Free PMC article. Review.
-
Proteins Recognizing DNA: Structural Uniqueness and Versatility of DNA-Binding Domains in Stem Cell Transcription Factors.Genes (Basel). 2017 Aug 1;8(8):192. doi: 10.3390/genes8080192. Genes (Basel). 2017. PMID: 28763006 Free PMC article. Review.
References
-
- Ambrosetti DC, Scholer HR, Dailey L, Basilico C (2000) Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem 275: 23387–23397 - PubMed
-
- Bogan AA, Thorn KS (1998) Anatomy of hot spots in protein interfaces. J Mol Biol 280: 1–9 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

