Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein
- PMID: 23878222
- PMCID: PMC3740854
- DOI: 10.1073/pnas.1305548110
Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein
Abstract
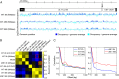
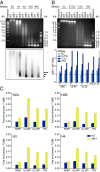
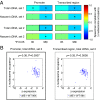
Human CMV (hCMV) establishes lifelong infections in most of us, causing developmental defects in human embryos and life-threatening disease in immunocompromised individuals. During productive infection, the viral >230,000-bp dsDNA genome is expressed widely and in a temporal cascade. The hCMV genome does not carry histones when encapsidated but has been proposed to form nucleosomes after release into the host cell nucleus. Here, we present hCMV genome-wide nucleosome occupancy and nascent transcript maps during infection of permissive human primary cells. We show that nucleosomes occupy nuclear viral DNA in a nonrandom and highly predictable fashion. At early times of infection, nucleosomes associate with the hCMV genome largely according to their intrinsic DNA sequence preferences, indicating that initial nucleosome formation is genetically encoded in the virus. However, as infection proceeds to the late phase, nucleosomes redistribute extensively to establish patterns mostly determined by nongenetic factors. We propose that these factors include key regulators of viral gene expression encoded at the hCMV major immediate-early (IE) locus. Indeed, mutant virus genomes deficient for IE1 expression exhibit globally increased nucleosome loads and reduced nucleosome dynamics compared with WT genomes. The temporal nucleosome occupancy differences between IE1-deficient and WT viruses correlate inversely with changes in the pattern of viral nascent and total transcript accumulation. These results provide a framework of spatial and temporal nucleosome organization across the genome of a major human pathogen and suggest that an hCMV major IE protein governs overall viral chromatin structure and function.
Keywords: ChIP-chip; epigenetic regulation; functional genomics; herpesvirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Human cytomegalovirus major immediate early 1 protein targets host chromosomes by docking to the acidic pocket on the nucleosome surface.J Virol. 2014 Jan;88(2):1228-48. doi: 10.1128/JVI.02606-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227840 Free PMC article.
-
Human Cytomegalovirus Genomes Survive Mitosis via the IE19 Chromatin-Tethering Domain.mBio. 2020 Sep 29;11(5):e02410-20. doi: 10.1128/mBio.02410-20. mBio. 2020. PMID: 32994332 Free PMC article.
-
Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells.J Virol. 2007 Sep;81(18):10123-36. doi: 10.1128/JVI.01009-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626080 Free PMC article.
-
How to control an infectious bead string: nucleosome-based regulation and targeting of herpesvirus chromatin.Rev Med Virol. 2011 May;21(3):154-80. doi: 10.1002/rmv.690. Rev Med Virol. 2011. PMID: 21538665 Review.
-
Chromatinisation of herpesvirus genomes.Rev Med Virol. 2010 Jan;20(1):34-50. doi: 10.1002/rmv.632. Rev Med Virol. 2010. PMID: 19890944 Review.
Cited by
-
Human Cytomegalovirus Infection Elicits Global Changes in Host Transcription by RNA Polymerases I, II, and III.Viruses. 2022 Apr 9;14(4):779. doi: 10.3390/v14040779. Viruses. 2022. PMID: 35458509 Free PMC article.
-
The 5' Untranslated Region of the Major Immediate Early mRNA Is Necessary for Efficient Human Cytomegalovirus Replication.J Virol. 2018 Mar 14;92(7):e02128-17. doi: 10.1128/JVI.02128-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343581 Free PMC article.
-
Cytomegalovirus latency and reactivation: recent insights into an age old problem.Rev Med Virol. 2016 Mar;26(2):75-89. doi: 10.1002/rmv.1862. Epub 2015 Nov 17. Rev Med Virol. 2016. PMID: 26572645 Free PMC article. Review.
-
Human cytomegalovirus and neonatal infection.Curr Res Microb Sci. 2024 Jun 24;7:100257. doi: 10.1016/j.crmicr.2024.100257. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39070527 Free PMC article. Review.
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
References
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319(5):1097–1113. - PubMed
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

