Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA)
- PMID: 23877245
- PMCID: PMC3794594
- DOI: 10.1093/nar/gkt634
Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA)
Abstract
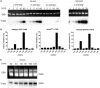
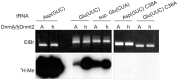
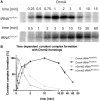
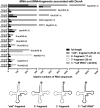
Although the DNA methyltransferase 2 family is highly conserved during evolution and recent reports suggested a dual specificity with stronger activity on transfer RNA (tRNA) than DNA substrates, the biological function is still obscure. We show that the Dictyostelium discoideum Dnmt2-homologue DnmA is an active tRNA methyltransferase that modifies C38 in tRNA(Asp(GUC)) in vitro and in vivo. By an ultraviolet-crosslinking and immunoprecipitation approach, we identified further DnmA targets. This revealed specific tRNA fragments bound by the enzyme and identified tRNA(Glu(CUC/UUC)) and tRNA(Gly(GCC)) as new but weaker substrates for both human Dnmt2 and DnmA in vitro but apparently not in vivo. Dnmt2 enzymes form transient covalent complexes with their substrates. The dynamics of complex formation and complex resolution reflect methylation efficiency in vitro. Quantitative PCR analyses revealed alterations in dnmA expression during development, cell cycle and in response to temperature stress. However, dnmA expression only partially correlated with tRNA methylation in vivo. Strikingly, dnmA expression in the laboratory strain AX2 was significantly lower than in the NC4 parent strain. As expression levels and binding of DnmA to a target in vivo are apparently not necessarily accompanied by methylation, we propose an additional biological function of DnmA apart from methylation.
Figures




Similar articles
-
The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu.Nucleic Acids Res. 2014 Jun;42(10):6487-96. doi: 10.1093/nar/gku256. Epub 2014 Apr 7. Nucleic Acids Res. 2014. PMID: 24711368 Free PMC article.
-
Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism.RNA. 2008 Aug;14(8):1663-70. doi: 10.1261/rna.970408. Epub 2008 Jun 20. RNA. 2008. PMID: 18567810 Free PMC article.
-
Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine.Nucleic Acids Res. 2015 Dec 15;43(22):10952-62. doi: 10.1093/nar/gkv980. Epub 2015 Sep 30. Nucleic Acids Res. 2015. PMID: 26424849 Free PMC article.
-
Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation.RNA Biol. 2017 Sep 2;14(9):1108-1123. doi: 10.1080/15476286.2016.1191737. Epub 2016 May 27. RNA Biol. 2017. PMID: 27232191 Free PMC article. Review.
-
Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification.Biomolecules. 2017 Feb 10;7(1):14. doi: 10.3390/biom7010014. Biomolecules. 2017. PMID: 28208632 Free PMC article. Review.
Cited by
-
NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation.EMBO J. 2016 Oct 4;35(19):2104-2119. doi: 10.15252/embj.201694885. Epub 2016 Aug 5. EMBO J. 2016. PMID: 27497299 Free PMC article.
-
An Optimized Microscale Thermophoresis Method for High-Throughput Screening of DNA Methyltransferase 2 Ligands.ACS Pharmacol Transl Sci. 2022 Oct 19;5(11):1079-1085. doi: 10.1021/acsptsci.2c00175. eCollection 2022 Nov 11. ACS Pharmacol Transl Sci. 2022. PMID: 36407957 Free PMC article.
-
Discovery of the major 15-30 nt mammalian small RNAs, their biogenesis and function.Nat Commun. 2023 Sep 18;14(1):5796. doi: 10.1038/s41467-023-41554-6. Nat Commun. 2023. PMID: 37723159 Free PMC article.
-
Ubiquitination-mediated degradation of TRDMT1 regulates homologous recombination and therapeutic response.NAR Cancer. 2021 Mar 22;3(1):zcab010. doi: 10.1093/narcan/zcab010. eCollection 2021 Mar. NAR Cancer. 2021. PMID: 33778494 Free PMC article.
-
Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum.Int J Mol Sci. 2020 Jul 23;21(15):5210. doi: 10.3390/ijms21155210. Int J Mol Sci. 2020. PMID: 32717856 Free PMC article.
References
-
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. - PubMed
-
- Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009;41:696–702. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

