Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium
- PMID: 23874508
- PMCID: PMC3709915
- DOI: 10.1371/journal.pone.0068095
Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium
Abstract
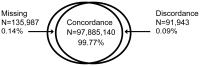
Genotyping arrays are a cost effective approach when typing previously-identified genetic polymorphisms in large numbers of samples. One limitation of genotyping arrays with rare variants (e.g., minor allele frequency [MAF] <0.01) is the difficulty that automated clustering algorithms have to accurately detect and assign genotype calls. Combining intensity data from large numbers of samples may increase the ability to accurately call the genotypes of rare variants. Approximately 62,000 ethnically diverse samples from eleven Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium cohorts were genotyped with the Illumina HumanExome BeadChip across seven genotyping centers. The raw data files for the samples were assembled into a single project for joint calling. To assess the quality of the joint calling, concordance of genotypes in a subset of individuals having both exome chip and exome sequence data was analyzed. After exclusion of low performing SNPs on the exome chip and non-overlap of SNPs derived from sequence data, genotypes of 185,119 variants (11,356 were monomorphic) were compared in 530 individuals that had whole exome sequence data. A total of 98,113,070 pairs of genotypes were tested and 99.77% were concordant, 0.14% had missing data, and 0.09% were discordant. We report that joint calling allows the ability to accurately genotype rare variation using array technology when large sample sizes are available and best practices are followed. The cluster file from this experiment is available at www.chargeconsortium.com/main/exomechip.
Conflict of interest statement
Figures
Similar articles
-
iCall: a genotype-calling algorithm for rare, low-frequency and common variants on the Illumina exome array.Bioinformatics. 2014 Jun 15;30(12):1714-20. doi: 10.1093/bioinformatics/btu107. Epub 2014 Feb 23. Bioinformatics. 2014. PMID: 24567545
-
Quality Control for the Illumina HumanExome BeadChip.Curr Protoc Hum Genet. 2016 Jul 1;90:2.14.1-2.14.16. doi: 10.1002/cphg.15. Curr Protoc Hum Genet. 2016. PMID: 27367164 Free PMC article.
-
Imputation-based assessment of next generation rare exome variant arrays.Pac Symp Biocomput. 2014:241-52. Pac Symp Biocomput. 2014. PMID: 24297551 Free PMC article.
-
A new strategy for enhancing imputation quality of rare variants from next-generation sequencing data via combining SNP and exome chip data.BMC Genomics. 2015 Dec 29;16:1109. doi: 10.1186/s12864-015-2192-y. BMC Genomics. 2015. PMID: 26715385 Free PMC article.
-
A Novel Quality-Control Procedure to Improve the Accuracy of Rare Variant Calling in SNP Arrays.Front Genet. 2021 Oct 26;12:736390. doi: 10.3389/fgene.2021.736390. eCollection 2021. Front Genet. 2021. PMID: 34764980 Free PMC article. Review.
Cited by
-
Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci.Nat Genet. 2016 Oct;48(10):1162-70. doi: 10.1038/ng.3660. Epub 2016 Sep 12. Nat Genet. 2016. PMID: 27618448 Free PMC article.
-
Identification of novel non-synonymous variants associated with type 2 diabetes-related metabolites in Korean population.Biosci Rep. 2019 Oct 30;39(10):BSR20190078. doi: 10.1042/BSR20190078. Biosci Rep. 2019. PMID: 31652446 Free PMC article.
-
Coding and regulatory variants are associated with serum protein levels and disease.Nat Commun. 2022 Jan 25;13(1):481. doi: 10.1038/s41467-022-28081-6. Nat Commun. 2022. PMID: 35079000 Free PMC article.
-
Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks.Am J Hum Genet. 2014 Feb 6;94(2):223-32. doi: 10.1016/j.ajhg.2014.01.009. Am J Hum Genet. 2014. PMID: 24507774 Free PMC article.
-
Frontotemporal dementia and its subtypes: a genome-wide association study.Lancet Neurol. 2014 Jul;13(7):686-99. doi: 10.1016/S1474-4422(14)70065-1. Lancet Neurol. 2014. PMID: 24943344 Free PMC article.
References
-
- Gudnason V, Sigurdsson G, Nissen H, Humphries SE (1997) Common founder mutation in the LDL receptor gene causing familial hypercholesterolaemia in the Icelandic population. Hum Mutat 10: 36–44. - PubMed
Publication types
MeSH terms
Grants and funding
- N01HC45205/HL/NHLBI NIH HHS/United States
- P30 DK035816/DK/NIDDK NIH HHS/United States
- N01 HC-95170/HC/NHLBI NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- HHSN268201000010C/HL/NHLBI NIH HHS/United States
- N01-HC-25195/HC/NHLBI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- U01 HL56565/HL/NHLBI NIH HHS/United States
- HHSN268201100001I/HL/NHLBI NIH HHS/United States
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- N01-HC-45205/HC/NHLBI NIH HHS/United States
- R01-HL-071258, ,/HL/NHLBI NIH HHS/United States
- N01-HC-95162/HC/NHLBI NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- N01HC45204/HC/NHLBI NIH HHS/United States
- N01AG12100/AG/NIA NIH HHS/United States
- N01-HC-05187/HC/NHLBI NIH HHS/United States
- N01 HC095170/HC/NHLBI NIH HHS/United States
- R01-HL-071252/HL/NHLBI NIH HHS/United States
- 5RC2HL102419/HL/NHLBI NIH HHS/United States
- R01 HL071251/HL/NHLBI NIH HHS/United States
- N01 HC-95171/HC/NHLBI NIH HHS/United States
- U01 HL56564/HL/NHLBI NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- R01 HL071259/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- HHSN2682011000010C/PHS HHS/United States
- UL1-RR-025005/RR/NCRR NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- N01 AG062101/AG/NIA NIH HHS/United States
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- N01-HC-48047/HC/NHLBI NIH HHS/United States
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- R01 HL071252/HL/NHLBI NIH HHS/United States
- N01HC48049/HL/NHLBI NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- HL105756/HL/NHLBI NIH HHS/United States
- R01-HL-071250/HL/NHLBI NIH HHS/United States
- N01 HC015103/HC/NHLBI NIH HHS/United States
- N01-HC-95168/HC/NHLBI NIH HHS/United States
- N01HC95169/HL/NHLBI NIH HHS/United States
- U01 HL056567/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- N01AG62101/AG/NIA NIH HHS/United States
- N01-HC-95163/HC/NHLBI NIH HHS/United States
- UL1-RR-024156/RR/NCRR NIH HHS/United States
- AG-15928/AG/NIA NIH HHS/United States
- R56 AG020098/AG/NIA NIH HHS/United States
- U01 HL56566/HL/NHLBI NIH HHS/United States
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- UL1 RR024156/RR/NCRR NIH HHS/United States
- N01-HC-45204/HC/NHLBI NIH HHS/United States
- R01 HL071250/HL/NHLBI NIH HHS/United States
- N01-HC-95159/HC/NHLBI NIH HHS/United States
- N01 HC095172/HC/NHLBI NIH HHS/United States
- 1R01AG032098-01A1/AG/NIA NIH HHS/United States
- R01 HL091244/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- DK-35816/DK/NIDDK NIH HHS/United States
- N01-HC-95095/HC/NHLBI NIH HHS/United States
- HHSN268201000011C/HL/NHLBI NIH HHS/United States
- N01-HC-95165/HC/NHLBI NIH HHS/United States
- HL087652/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- N01 AG062106/AG/NIA NIH HHS/United States
- U01 HL56569/HL/NHLBI NIH HHS/United States
- R01-HL-071251/HL/NHLBI NIH HHS/United States
- N01HC55222/HL/NHLBI NIH HHS/United States
- R01-HL-084099/HL/NHLBI NIH HHS/United States
- N01-HC-85086/HC/NHLBI NIH HHS/United States
- N01 HC095171/HC/NHLBI NIH HHS/United States
- N01HC85086/HL/NHLBI NIH HHS/United States
- AG-027058/AG/NIA NIH HHS/United States
- R01 AG032098/AG/NIA NIH HHS/United States
- N01-HC-48050/HC/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- T32 HL007208/HL/NHLBI NIH HHS/United States
- U01 HG004729/HG/NHGRI NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- N01-HC-35129/HC/NHLBI NIH HHS/United States
- N01-HC-45134/HC/NHLBI NIH HHS/United States
- N01 HC-55222/HC/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 HL071051/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HL41993/HL/NHLBI NIH HHS/United States
- R01-HL-071259/HL/NHLBI NIH HHS/United States
- N01HC95095/HL/NHLBI NIH HHS/United States
- N01-HC-95169/HC/NHLBI NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- N01 AG062103/AG/NIA NIH HHS/United States
- N01-HC-95164/HC/NHLBI NIH HHS/United States
- N01HC95159/HL/NHLBI NIH HHS/United States
- R01-HL-071205/HL/NHLBI NIH HHS/United States
- N01-HC-48049/HC/NHLBI NIH HHS/United States
- N01 HC-95172/HC/NHLBI NIH HHS/United States
- N01-HC-75150/HC/NHLBI NIH HHS/United States
- R01 HL084099/HL/NHLBI NIH HHS/United States
- N01 HC045134/HC/NHLBI NIH HHS/United States
- HHSN268201100002C/WH/WHI NIH HHS/United States
- HHSN2682011000012C/PHS HHS/United States
- N01-HC-95160/HC/NHLBI NIH HHS/United States
- N01 HC-15103/HC/NHLBI NIH HHS/United States
- R01 HL080295/HL/NHLBI NIH HHS/United States
- U01 HL56568/HL/NHLBI NIH HHS/United States
- R01 AG020098/AG/NIA NIH HHS/United States
- U01-HG-004729/HG/NHGRI NIH HHS/United States
- N01HC48050/HL/NHLBI NIH HHS/United States
- K01 HL070444/HL/NHLBI NIH HHS/United States
- N01HC75150/HL/NHLBI NIH HHS/United States
- N01HC05187/HL/NHLBI NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- N01HC48047/HL/NHLBI NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- N01HC25195/HL/NHLBI NIH HHS/United States
- R01 HL071205/HL/NHLBI NIH HHS/United States
- DK063491/DK/NIDDK NIH HHS/United States
- U01 HL56567/HL/NHLBI NIH HHS/United States
- N01-AG-12100/AG/NIA NIH HHS/United States
- N01-HC-45133/HC/NHLBI NIH HHS/United States
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- K01-HL70444/HL/NHLBI NIH HHS/United States
- N01AG62103/AG/NIA NIH HHS/United States
- N01-HC-85079/HC/NHLBI NIH HHS/United States
- N01-HC-95161/HC/NHLBI NIH HHS/United States
- HHSN268201100002I/HL/NHLBI NIH HHS/United States
- U01 HL56563/HL/NHLBI NIH HHS/United States
- HL080295/HL/NHLBI NIH HHS/United States
- N01AG62106/AG/NIA NIH HHS/United States
- HHSN268201000012C/HL/NHLBI NIH HHS/United States
- N01-HC-85239/HC/NHLBI NIH HHS/United States
- N01-HC-48048/HC/NHLBI NIH HHS/United States
- AG-023629/AG/NIA NIH HHS/United States
- N01HC85079/HL/NHLBI NIH HHS/United States
- R01- HL-071051/HL/NHLBI NIH HHS/United States
- N01-HC-95166/HC/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 HL041993/HL/NHLBI NIH HHS/United States
- R01 AG027058/AG/NIA NIH HHS/United States
- N01 HC045133/HC/NHLBI NIH HHS/United States
- HL091244/HL/NHLBI NIH HHS/United States
- R01 HL071258/HL/NHLBI NIH HHS/United States
- HHSN268201100001C/WH/WHI NIH HHS/United States
- N01 HC035129/HC/NHLBI NIH HHS/United States
- R56 AG023629/AG/NIA NIH HHS/United States
- N01HC48048/HL/NHLBI NIH HHS/United States
- N01-HC-95167/HC/NHLBI NIH HHS/United States
- HHSN2682011000011C/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous