Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration
- PMID: 23871589
- PMCID: PMC3873200
- DOI: 10.1016/j.devcel.2013.06.017
Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration
Abstract
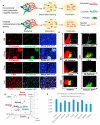
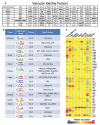
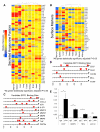
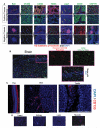
Microvascular endothelial cells (ECs) within different tissues are endowed with distinct but as yet unrecognized structural, phenotypic, and functional attributes. We devised EC purification, cultivation, profiling, and transplantation models that establish tissue-specific molecular libraries of ECs devoid of lymphatic ECs or parenchymal cells. These libraries identify attributes that confer ECs with their organotypic features. We show that clusters of transcription factors, angiocrine growth factors, adhesion molecules, and chemokines are expressed in unique combinations by ECs of each organ. Furthermore, ECs respond distinctly in tissue regeneration models, hepatectomy, and myeloablation. To test the data set, we developed a transplantation model that employs generic ECs differentiated from embryonic stem cells. Transplanted generic ECs engraft into regenerating tissues and acquire features of organotypic ECs. Collectively, we demonstrate the utility of informational databases of ECs toward uncovering the extravascular and intrinsic signals that define EC heterogeneity. These factors could be exploited therapeutically to engineer tissue-specific ECs for regeneration.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Endothelial heterogeneity and their relevance in cardiac development and coronary artery disease.Vascul Pharmacol. 2023 Dec;153:107242. doi: 10.1016/j.vph.2023.107242. Epub 2023 Nov 7. Vascul Pharmacol. 2023. PMID: 37940065 Review.
-
ROCK suppression promotes differentiation and expansion of endothelial cells from embryonic stem cell-derived Flk1(+) mesodermal precursor cells.Blood. 2012 Sep 27;120(13):2733-44. doi: 10.1182/blood-2012-04-421610. Epub 2012 Aug 14. Blood. 2012. PMID: 22896004
-
Developmental angiocrine diversification of endothelial cells for organotypic regeneration.Dev Cell. 2021 Nov 22;56(22):3042-3051. doi: 10.1016/j.devcel.2021.10.020. Dev Cell. 2021. PMID: 34813766 Review.
-
Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA.Cell Res. 2013 Oct;23(10):1172-86. doi: 10.1038/cr.2013.112. Epub 2013 Sep 10. Cell Res. 2013. PMID: 24018375 Free PMC article.
-
Reversal of endothelial progenitor cell dysfunction in patients with type 2 diabetes using a conditioned medium of human embryonic stem cell-derived endothelial cells.Diabetes Metab Res Rev. 2012 Jul;28(5):462-73. doi: 10.1002/dmrr.2304. Epub 2012 Apr 10. Diabetes Metab Res Rev. 2012. PMID: 22492468
Cited by
-
Endothelial JAK2V617F mutation leads to thrombosis, vasculopathy, and cardiomyopathy in a murine model of myeloproliferative neoplasm.J Thromb Haemost. 2020 Dec;18(12):3359-3370. doi: 10.1111/jth.15095. Epub 2020 Oct 5. J Thromb Haemost. 2020. PMID: 32920974 Free PMC article.
-
Toward a granular molecular-anatomic map of the blood vasculature - single-cell RNA sequencing makes the leap.Ups J Med Sci. 2022 Oct 21;127. doi: 10.48101/ujms.v127.9051. eCollection 2022. Ups J Med Sci. 2022. PMID: 36337278 Free PMC article. Review.
-
A Novel Experimental Approach for In Vivo Analyses of the Salivary Gland Microvasculature.Front Immunol. 2021 Feb 17;11:604470. doi: 10.3389/fimmu.2020.604470. eCollection 2020. Front Immunol. 2021. PMID: 33679695 Free PMC article.
-
Pancreatic cancer cell-derived exosomal microRNA-27a promotes angiogenesis of human microvascular endothelial cells in pancreatic cancer via BTG2.J Cell Mol Med. 2020 Jan;24(1):588-604. doi: 10.1111/jcmm.14766. Epub 2019 Nov 13. J Cell Mol Med. 2020. PMID: 31724333 Free PMC article.
-
Endothelial Cell Development and Its Application to Regenerative Medicine.Circ Res. 2019 Aug 2;125(4):489-501. doi: 10.1161/CIRCRESAHA.119.311405. Epub 2019 Aug 1. Circ Res. 2019. PMID: 31518171 Free PMC article. Review.
References
-
- Ahn J, Ko M, Lee K, Oh J, Jeon SH, Seong RH. Expression of SRG3, a core component of mouse SWI/SNF chromatin-remodeling complex, is regulated by cooperative interactions between Sp1/Sp3 and Ets transcription factors. Biochem. Biophys. Res. Commun. 2005;338:1435–1446. - PubMed
-
- Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 2007;100:158–173. - PubMed
-
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn. 2006;235:759–767. - PubMed
-
- Børsum T, Hagen I, Henriksen T, Carlander B. Alterations in the protein composition and surface structure of human endothelial cells during growth in primary culture. Atherosclerosis. 1982;44:367–378. - PubMed
-
- Brauer PR, Cai DH. Expression of tissue inhibitor of metalloproteinases (TIMPs) during early cardiac development. Mech. Dev. 2002;113:175–179. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

