NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers
- PMID: 23868852
- PMCID: PMC3740838
- DOI: 10.1073/pnas.1305688110
NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers
Abstract
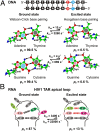
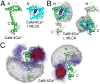
The importance of dynamics to biomolecular function is becoming increasingly clear. A description of the structure-function relationship must, therefore, include the role of motion, requiring a shift in paradigm from focus on a single static 3D picture to one where a given biomolecule is considered in terms of an ensemble of interconverting conformers, each with potentially diverse activities. In this Perspective, we describe how recent developments in solution NMR spectroscopy facilitate atomic resolution studies of sparsely populated, transiently formed biomolecular conformations that exchange with the native state. Examples of how this methodology is applied to protein folding and misfolding, ligand binding, and molecular recognition are provided as a means of illustrating both the power of the new techniques and the significant roles that conformationally excited protein states play in biology.
Keywords: energy landscape; invisible states; structure–function paradigm; transiently and sparsely populated biomolecular states.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Visualizing side chains of invisible protein conformers by solution NMR.J Mol Biol. 2014 Feb 6;426(3):763-74. doi: 10.1016/j.jmb.2013.10.041. Epub 2013 Nov 8. J Mol Biol. 2014. PMID: 24211467
-
Measuring hydrogen exchange rates in invisible protein excited states.Proc Natl Acad Sci U S A. 2014 Jun 17;111(24):8820-5. doi: 10.1073/pnas.1405011111. Epub 2014 Jun 2. Proc Natl Acad Sci U S A. 2014. PMID: 24889628 Free PMC article.
-
Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding.Acc Chem Res. 2008 Mar;41(3):442-51. doi: 10.1021/ar700189y. Epub 2008 Feb 15. Acc Chem Res. 2008. PMID: 18275162 Review.
-
Unveiling invisible protein states with NMR spectroscopy.Curr Opin Struct Biol. 2020 Feb;60:39-49. doi: 10.1016/j.sbi.2019.10.008. Epub 2019 Dec 11. Curr Opin Struct Biol. 2020. PMID: 31835059 Review.
-
Measurement of methyl group motional parameters of invisible, excited protein states by NMR spectroscopy.J Am Chem Soc. 2009 Sep 9;131(35):12745-54. doi: 10.1021/ja903897e. J Am Chem Soc. 2009. PMID: 19685870
Cited by
-
2'-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states.Nucleic Acids Res. 2020 Dec 2;48(21):12365-12379. doi: 10.1093/nar/gkaa928. Nucleic Acids Res. 2020. PMID: 33104789 Free PMC article.
-
Rational design of hairpin RNA excited states reveals multi-step transitions.Nat Commun. 2022 Mar 21;13(1):1523. doi: 10.1038/s41467-022-29194-8. Nat Commun. 2022. PMID: 35314698 Free PMC article.
-
Chemical shift-based methods in NMR structure determination.Prog Nucl Magn Reson Spectrosc. 2018 Jun-Aug;106-107:1-25. doi: 10.1016/j.pnmrs.2018.03.002. Epub 2018 Mar 11. Prog Nucl Magn Reson Spectrosc. 2018. PMID: 31047599 Free PMC article. Review.
-
Biophysics of protein evolution and evolutionary protein biophysics.J R Soc Interface. 2014 Nov 6;11(100):20140419. doi: 10.1098/rsif.2014.0419. J R Soc Interface. 2014. PMID: 25165599 Free PMC article.
-
Crystal cryocooling distorts conformational heterogeneity in a model Michaelis complex of DHFR.Structure. 2014 Jun 10;22(6):899-910. doi: 10.1016/j.str.2014.04.016. Epub 2014 May 29. Structure. 2014. PMID: 24882744 Free PMC article.
References
-
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. - PubMed
-
- Kendrew JC, et al. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181(4610):662–666. - PubMed
-
- Perutz MF, et al. Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185(4711):416–422. - PubMed
-
- Blake CC, et al. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206(4986):757–761. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

