Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4
- PMID: 23862699
- PMCID: PMC3756530
- DOI: 10.1021/bi400684q
Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4
Abstract
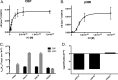
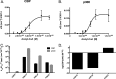
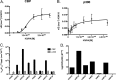
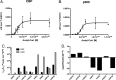
Although p300 and CBP lysine acetyltransferases are often treated interchangeably, the inability of one enzyme to compensate for the loss of the other suggests unique roles for each. As these deficiencies coincide with aberrant levels of histone acetylation, we hypothesized that the key difference between p300 and CBP activity is differences in their specificity/selectivity for lysines within the histones. Utilizing a label-free, quantitative mass spectrometry based technique, we determined the kinetic parameters of both CBP and p300 at each lysine of H3 and H4, under conditions we would expect to encounter in the cell (either limiting acetyl-CoA or histone). Our results show that while p300 and CBP acetylate many common residues on H3 and H4, they do in fact possess very different specificities, and these specificities are dependent on whether histone or acetyl-CoA is limiting. Steady-state experiments with limiting H3 demonstrate that both CBP and p300 acetylate H3K14, H3K18, H3K23, with p300 having specificities up to 10¹⁰-fold higher than CBP. Utilizing tetramer as a substrate, both enzymes also acetylate H4K5, H4K8, H4K12, and H4K16. With limiting tetramer, CBP displays higher specificities, especially at H3K18, where CBP specificity is 10³²-fold higher than p300. With limiting acetyl-CoA, p300 has the highest specificity at H4K16, where specificity is 10¹⁸-fold higher than CBP. This discovery of unique specificity for targets of CBP- vs p300-mediated acetylation of histone lysine residues presents a new model for understanding their respective biological roles and possibly an opportunity for selective therapeutic intervention.
Figures
Similar articles
-
Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation.EMBO J. 2011 Jan 19;30(2):249-62. doi: 10.1038/emboj.2010.318. Epub 2010 Dec 3. EMBO J. 2011. PMID: 21131905 Free PMC article.
-
Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3.PLoS One. 2013;8(2):e54896. doi: 10.1371/journal.pone.0054896. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437046 Free PMC article.
-
Effect of melatonin on neuronal differentiation requires CBP/p300-mediated acetylation of histone H3 lysine 14.Neuroscience. 2017 Nov 19;364:45-59. doi: 10.1016/j.neuroscience.2017.07.064. Epub 2017 Aug 4. Neuroscience. 2017. PMID: 28782640
-
Defining the orphan functions of lysine acetyltransferases.ACS Chem Biol. 2015 Jan 16;10(1):85-94. doi: 10.1021/cb500853p. ACS Chem Biol. 2015. PMID: 25591746 Free PMC article. Review.
-
CREBBP and p300 lysine acetyl transferases in the DNA damage response.Cell Mol Life Sci. 2018 Apr;75(8):1325-1338. doi: 10.1007/s00018-017-2717-4. Epub 2017 Nov 23. Cell Mol Life Sci. 2018. PMID: 29170789 Free PMC article. Review.
Cited by
-
Human EHMT2/G9a activates p53 through methylation-independent mechanism.Oncogene. 2017 Feb 16;36(7):922-932. doi: 10.1038/onc.2016.258. Epub 2016 Jul 25. Oncogene. 2017. PMID: 27452519
-
Paradoxical activation of AMPK by glucose drives selective EP300 activity in colorectal cancer.PLoS Biol. 2020 Jun 30;18(6):e3000732. doi: 10.1371/journal.pbio.3000732. eCollection 2020 Jun. PLoS Biol. 2020. PMID: 32603375 Free PMC article.
-
Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation.J Exp Clin Cancer Res. 2019 Jun 13;38(1):251. doi: 10.1186/s13046-019-1242-8. J Exp Clin Cancer Res. 2019. PMID: 31196146 Free PMC article.
-
Acetylation of STX17 (syntaxin 17) controls autophagosome maturation.Autophagy. 2021 May;17(5):1157-1169. doi: 10.1080/15548627.2020.1752471. Epub 2020 Apr 15. Autophagy. 2021. PMID: 32264736 Free PMC article.
-
New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids.J Biol Chem. 2014 May 30;289(22):15798-809. doi: 10.1074/jbc.M113.544346. Epub 2014 Apr 23. J Biol Chem. 2014. PMID: 24759103 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

