Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells
- PMID: 23861660
- PMCID: PMC3701696
- DOI: 10.1371/journal.pcbi.1003122
Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells
Abstract
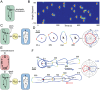
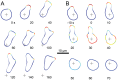
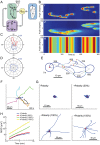
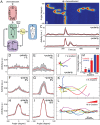
Chemotaxis involves the coordinated action of separable but interrelated processes: motility, gradient sensing, and polarization. We have hypothesized that these are mediated by separate modules that account for these processes individually and that, when combined, recreate most of the behaviors of chemotactic cells. Here, we describe a mathematical model where the modules are implemented in terms of reaction-diffusion equations. Migration and the accompanying changes in cellular morphology are demonstrated in simulations using a mechanical model of the cell cortex implemented in the level set framework. The central module is an excitable network that accounts for random migration. The response to combinations of uniform stimuli and gradients is mediated by a local excitation, global inhibition module that biases the direction in which excitability is directed. A polarization module linked to the excitable network through the cytoskeleton allows unstimulated cells to move persistently and, for cells in gradients, to gradually acquire distinct sensitivity between front and back. Finally, by varying the strengths of various feedback loops in the model we obtain cellular behaviors that mirror those of genetically altered cell lines.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity.Annu Rev Biophys. 2010;39:265-89. doi: 10.1146/annurev.biophys.093008.131228. Annu Rev Biophys. 2010. PMID: 20192768 Free PMC article. Review.
-
Cells navigate with a local-excitation, global-inhibition-biased excitable network.Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17079-86. doi: 10.1073/pnas.1011271107. Epub 2010 Sep 23. Proc Natl Acad Sci U S A. 2010. PMID: 20864631 Free PMC article.
-
Spontaneous signal generation by an excitable system for cell migration.Front Cell Dev Biol. 2024 Feb 28;12:1373609. doi: 10.3389/fcell.2024.1373609. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38481533 Free PMC article. Review.
-
A conceptual molecular network for chemotactic behaviors characterized by feedback of molecules cycling between the membrane and the cytosol.Sci Signal. 2010 Dec 14;3(152):ra89. doi: 10.1126/scisignal.2001056. Sci Signal. 2010. PMID: 21156936
-
Three-dimensional stochastic simulation of chemoattractant-mediated excitability in cells.PLoS Comput Biol. 2021 Jul 14;17(7):e1008803. doi: 10.1371/journal.pcbi.1008803. eCollection 2021 Jul. PLoS Comput Biol. 2021. PMID: 34260581 Free PMC article.
Cited by
-
Spatiotemporal dynamics of membrane surface charge regulates cell polarity and migration.Nat Cell Biol. 2022 Oct;24(10):1499-1515. doi: 10.1038/s41556-022-00997-7. Epub 2022 Oct 6. Nat Cell Biol. 2022. PMID: 36202973 Free PMC article.
-
Progress and perspectives in signal transduction, actin dynamics, and movement at the cell and tissue level: lessons from Dictyostelium.Interface Focus. 2016 Oct 6;6(5):20160047. doi: 10.1098/rsfs.2016.0047. Interface Focus. 2016. PMID: 27708767 Free PMC article. Review.
-
Anomalous diffusion and asymmetric tempering memory in neutrophil chemotaxis.PLoS Comput Biol. 2022 May 18;18(5):e1010089. doi: 10.1371/journal.pcbi.1010089. eCollection 2022 May. PLoS Comput Biol. 2022. PMID: 35584137 Free PMC article.
-
Co-existence of Ras activation in a chemotactic signal transduction pathway and in an autonomous wave - forming system.Small GTPases. 2019 Jan;10(1):72-80. doi: 10.1080/21541248.2016.1268666. Epub 2017 Jan 31. Small GTPases. 2019. PMID: 28136018 Free PMC article.
-
Coupled excitable Ras and F-actin activation mediates spontaneous pseudopod formation and directed cell movement.Mol Biol Cell. 2017 Apr 1;28(7):922-934. doi: 10.1091/mbc.E16-10-0733. Epub 2017 Feb 1. Mol Biol Cell. 2017. PMID: 28148648 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

