The human cytomegalovirus US27 gene product enhances cell proliferation and alters cellular gene expression
- PMID: 23850869
- PMCID: PMC3774267
- DOI: 10.1016/j.virusres.2013.07.002
The human cytomegalovirus US27 gene product enhances cell proliferation and alters cellular gene expression
Abstract
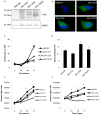
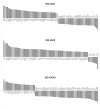
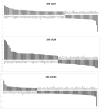
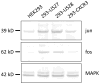
Human cytomegalovirus (HCMV) is a prevalent pathogen worldwide. Although generally harmless in healthy individuals, HCMV can pose a serious threat to immune compromised individuals and developing fetuses in utero. HCMV encodes four genes predicted to give rise to G protein-coupled receptors (GPCRs): US27, US28, UL33, and UL78. The US28 gene product is a functional chemokine receptor that enhances cell growth in some cell types but induces apoptosis in others. In contrast, the US27 gene product has not been demonstrated to signal either constitutively or in a ligand-induced manner. In this study, US27 was expressed in transfected cells, and both cell proliferation and DNA synthesis were significantly increased compared to control cells. PCR array analysis revealed that expression of US27 led to changes in a limited number of cellular genes, but genes that were up-regulated included the pro-survival factor Bcl-x, AP-1 transcription factor components jun and fos, and the IL-6 family cytokine oncostatin M. These results demonstrate that US27 can impact host cell physiology and may shed light on the function of this orphan viral GPCR.
Keywords: Chemokine receptor; Cytomegalovirus; GPCR; HCMV.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures

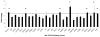
Similar articles
-
The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through Gβγ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1.J Virol. 2018 Nov 12;92(23):e00644-18. doi: 10.1128/JVI.00644-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209167 Free PMC article.
-
The DRY box and C-terminal domain of the human cytomegalovirus US27 gene product play a role in promoting cell growth and survival.PLoS One. 2014 Nov 19;9(11):e113427. doi: 10.1371/journal.pone.0113427. eCollection 2014. PLoS One. 2014. PMID: 25409008 Free PMC article.
-
Evolution of the ability to modulate host chemokine networks via gene duplication in human cytomegalovirus (HCMV).Infect Genet Evol. 2017 Jul;51:46-53. doi: 10.1016/j.meegid.2017.03.013. Epub 2017 Mar 14. Infect Genet Evol. 2017. PMID: 28315475 Free PMC article.
-
The human cytomegalovirus chemokine receptor homolog encoded by US27.Virus Genes. 2017 Aug;53(4):516-521. doi: 10.1007/s11262-017-1462-y. Epub 2017 Apr 26. Virus Genes. 2017. PMID: 28447191 Review.
-
Emerging roles of cytomegalovirus-encoded G protein-coupled receptors during lytic and latent infection.Med Microbiol Immunol. 2019 Aug;208(3-4):447-456. doi: 10.1007/s00430-019-00595-9. Epub 2019 Mar 21. Med Microbiol Immunol. 2019. PMID: 30900091 Review.
Cited by
-
Human Cytomegalovirus Primary Infection and Reactivation: Insights From Virion-Carried Molecules.Front Microbiol. 2020 Jul 14;11:1511. doi: 10.3389/fmicb.2020.01511. eCollection 2020. Front Microbiol. 2020. PMID: 32765441 Free PMC article. Review.
-
Herpesvirus-encoded GPCRs: neglected players in inflammatory and proliferative diseases?Nat Rev Drug Discov. 2014 Feb;13(2):123-39. doi: 10.1038/nrd4189. Epub 2014 Jan 21. Nat Rev Drug Discov. 2014. PMID: 24445563 Review.
-
Viral manipulation of the host immune response.Curr Opin Immunol. 2015 Oct;36:54-60. doi: 10.1016/j.coi.2015.06.012. Epub 2015 Jul 10. Curr Opin Immunol. 2015. PMID: 26177523 Free PMC article. Review.
-
The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through Gβγ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1.J Virol. 2018 Nov 12;92(23):e00644-18. doi: 10.1128/JVI.00644-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209167 Free PMC article.
-
Virion Glycoprotein-Mediated Immune Evasion by Human Cytomegalovirus: a Sticky Virus Makes a Slick Getaway.Microbiol Mol Biol Rev. 2016 Jun 15;80(3):663-77. doi: 10.1128/MMBR.00018-16. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27307580 Free PMC article. Review.
References
-
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072(2–3):129–157. - PubMed
-
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gershengorn MC, Mesri EA. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391(6662):86–89. - PubMed
-
- Beisser PS, Goh CS, Cohen FE, Michelson S. Viral chemokine receptors and chemokines in human cytomegalovirus trafficking and interaction with the immune system. CMV chemokine receptors. Curr Top Microbiol Immunol. 2002;269:203–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

