Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion
- PMID: 23836939
- PMCID: PMC3783167
- DOI: 10.1093/nar/gkt584
Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion
Abstract
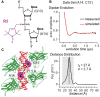
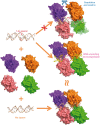
The p53 core domain binds to response elements (REs) that contain two continuous half-sites as a cooperative tetramer, but how p53 recognizes discontinuous REs is not well understood. Here we describe the crystal structure of the p53 core domain bound to a naturally occurring RE located at the promoter of the Bcl-2-associated X protein (BAX) gene, which contains a one base-pair insertion between the two half-sites. Surprisingly, p53 forms a tetramer on the BAX-RE that is nearly identical to what has been reported on other REs with a 0-bp spacer. Each p53 dimer of the tetramer binds in register to a half-site and maintains the same protein-DNA interactions as previously observed, and the two dimers retain all the protein-protein contacts without undergoing rotation or translation. To accommodate the additional base pair, the DNA is deformed and partially disordered around the spacer region, resulting in an apparent unwinding and compression, such that the interactions between the dimers are maintained. Furthermore, DNA deformation within the p53-bound BAX-RE is confirmed in solution by site-directed spin labeling measurements. Our results provide a structural insight into the mechanism by which p53 binds to discontinuous sites with one base-pair spacer.
Figures



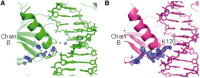
Similar articles
-
Diverse p53/DNA binding modes expand the repertoire of p53 response elements.Proc Natl Acad Sci U S A. 2017 Oct 3;114(40):10624-10629. doi: 10.1073/pnas.1618005114. Epub 2017 Sep 14. Proc Natl Acad Sci U S A. 2017. PMID: 28912355 Free PMC article.
-
Conformations of p53 response elements in solution deduced using site-directed spin labeling and Monte Carlo sampling.Nucleic Acids Res. 2014 Feb;42(4):2789-97. doi: 10.1093/nar/gkt1219. Epub 2013 Nov 30. Nucleic Acids Res. 2014. PMID: 24293651 Free PMC article.
-
Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer.Structure. 2010 Feb 10;18(2):246-56. doi: 10.1016/j.str.2009.11.011. Structure. 2010. PMID: 20159469 Free PMC article.
-
Roles of p53 Family Structure and Function in Non-Canonical Response Element Binding and Activation.Int J Mol Sci. 2019 Jul 27;20(15):3681. doi: 10.3390/ijms20153681. Int J Mol Sci. 2019. PMID: 31357595 Free PMC article. Review.
-
The Rich World of p53 DNA Binding Targets: The Role of DNA Structure.Int J Mol Sci. 2019 Nov 9;20(22):5605. doi: 10.3390/ijms20225605. Int J Mol Sci. 2019. PMID: 31717504 Free PMC article. Review.
Cited by
-
Expanding Roles of the E2F-RB-p53 Pathway in Tumor Suppression.Biology (Basel). 2023 Dec 11;12(12):1511. doi: 10.3390/biology12121511. Biology (Basel). 2023. PMID: 38132337 Free PMC article. Review.
-
DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.Nucleic Acids Res. 2015 Jan;43(2):1268-82. doi: 10.1093/nar/gku1373. Epub 2015 Jan 7. Nucleic Acids Res. 2015. PMID: 25567984 Free PMC article.
-
Protein-induced DNA linking number change by sequence-specific DNA binding proteins and its biological effects.Biophys Rev. 2016 Sep;8(3):197-207. doi: 10.1007/s12551-016-0204-z. Epub 2016 Jun 10. Biophys Rev. 2016. PMID: 28510223 Free PMC article. Review.
-
Cell proliferation activity delineated by molecular docking of four new compounds isolated from the aerial parts of Suaeda monoica Forssk. ex. J.F. Gmel.Saudi Pharm J. 2020 Feb;28(2):172-186. doi: 10.1016/j.jsps.2019.11.019. Epub 2019 Dec 7. Saudi Pharm J. 2020. PMID: 32042256 Free PMC article.
-
Gastric Inhibitory Polypeptide Receptor (GIPR) Overexpression Reduces the Tumorigenic Potential of Retinoblastoma Cells.Cancers (Basel). 2024 Apr 25;16(9):1656. doi: 10.3390/cancers16091656. Cancers (Basel). 2024. PMID: 38730608 Free PMC article.
References
-
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. - PubMed
-
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. - PubMed
-
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. - PubMed
-
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

