Something silent this way forms: the functional organization of the repressive nuclear compartment
- PMID: 23834025
- PMCID: PMC3999972
- DOI: 10.1146/annurev-cellbio-101512-122317
Something silent this way forms: the functional organization of the repressive nuclear compartment
Abstract
The repressive compartment of the nucleus is comprised primarily of telomeric and centromeric regions, the silent portion of ribosomal RNA genes, the majority of transposable element repeats, and facultatively repressed genes specific to different cell types. This compartment localizes into three main regions: the peripheral heterochromatin, perinucleolar heterochromatin, and pericentromeric heterochromatin. Both chromatin remodeling proteins and transcription of noncoding RNAs are involved in maintenance of repression in these compartments. Global reorganization of the repressive compartment occurs at each cell division, during early development, and during terminal differentiation. Differential action of chromatin remodeling complexes and boundary element looping activities are involved in mediating these organizational changes. We discuss the evidence that heterochromatin formation and compartmentalization may drive nuclear organization.
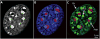
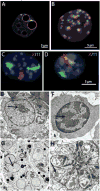
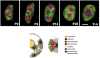
Figures



Similar articles
-
Functional redundancy in the nuclear compartmentalization of the late-replicating genome.Nucleus. 2014;5(6):626-35. doi: 10.4161/19491034.2014.990863. Nucleus. 2014. PMID: 25493640 Free PMC article.
-
PfMORC protein regulates chromatin accessibility and transcriptional repression in the human malaria parasite, Plasmodium falciparum.Elife. 2024 Dec 5;12:RP92499. doi: 10.7554/eLife.92499. Elife. 2024. PMID: 39636094 Free PMC article.
-
Prevalent fast evolution of genes involved in heterochromatin functions.bioRxiv [Preprint]. 2024 Jul 23:2024.03.03.583199. doi: 10.1101/2024.03.03.583199. bioRxiv. 2024. Update in: Mol Biol Evol. 2024 Sep 4;41(9):msae181. doi: 10.1093/molbev/msae181. PMID: 38496614 Free PMC article. Updated. Preprint.
-
Impact of nuclear organization and dynamics on epigenetic regulation in the central nervous system: implications for neurological disease states.Ann N Y Acad Sci. 2010 Sep;1204 Suppl(Suppl):E20-37. doi: 10.1111/j.1749-6632.2010.05718.x. Ann N Y Acad Sci. 2010. PMID: 20840166 Free PMC article. Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
Cited by
-
Nucleolar tethering mediates pairing between the IgH and Myc loci.Nucleus. 2014 Sep-Oct;5(5):474-81. doi: 10.4161/nucl.36233. Nucleus. 2014. PMID: 25482199 Free PMC article.
-
Effects of Chromatin Structure Modifiers on the trans-Acting Heterochromatin Position Effect in Drosophila melanogaster.Dokl Biochem Biophys. 2023 Dec;513(Suppl 1):S75-S81. doi: 10.1134/S160767292470073X. Epub 2024 Feb 20. Dokl Biochem Biophys. 2023. PMID: 38379078
-
82-kDa choline acetyltransferase and SATB1 localize to β-amyloid induced matrix attachment regions.Sci Rep. 2016 Apr 7;6:23914. doi: 10.1038/srep23914. Sci Rep. 2016. PMID: 27052102 Free PMC article.
-
Keep moving and stay in a good shape to find your homologous recombination partner.Curr Genet. 2019 Feb;65(1):29-39. doi: 10.1007/s00294-018-0873-1. Epub 2018 Aug 10. Curr Genet. 2019. PMID: 30097675 Free PMC article. Review.
-
The PRR14 heterochromatin tether encodes modular domains that mediate and regulate nuclear lamina targeting.J Cell Sci. 2020 May 27;133(10):jcs240416. doi: 10.1242/jcs.240416. J Cell Sci. 2020. PMID: 32317397 Free PMC article.
References
-
- Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–71. - PubMed
-
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–90. - PubMed
-
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

