The mean and variance of phylogenetic diversity under rarefaction
- PMID: 23833701
- PMCID: PMC3699894
- DOI: 10.1111/2041-210X.12042
The mean and variance of phylogenetic diversity under rarefaction
Abstract
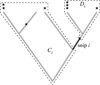
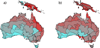
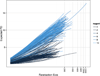
Phylogenetic diversity (PD) depends on sampling depth, which complicates the comparison of PD between samples of different depth. One approach to dealing with differing sample depth for a given diversity statistic is to rarefy, which means to take a random subset of a given size of the original sample. Exact analytical formulae for the mean and variance of species richness under rarefaction have existed for some time but no such solution exists for PD.We have derived exact formulae for the mean and variance of PD under rarefaction. We confirm that these formulae are correct by comparing exact solution mean and variance to that calculated by repeated random (Monte Carlo) subsampling of a dataset of stem counts of woody shrubs of Toohey Forest, Queensland, Australia. We also demonstrate the application of the method using two examples: identifying hotspots of mammalian diversity in Australasian ecoregions, and characterising the human vaginal microbiome.There is a very high degree of correspondence between the analytical and random subsampling methods for calculating mean and variance of PD under rarefaction, although the Monte Carlo method requires a large number of random draws to converge on the exact solution for the variance.Rarefaction of mammalian PD of ecoregions in Australasia to a common standard of 25 species reveals very different rank orderings of ecoregions, indicating quite different hotspots of diversity than those obtained for unrarefied PD. The application of these methods to the vaginal microbiome shows that a classical score used to quantify bacterial vaginosis is correlated with the shape of the rarefaction curve.The analytical formulae for the mean and variance of PD under rarefaction are both exact and more efficient than repeated subsampling. Rarefaction of PD allows for many applications where comparisons of samples of different depth is required.
Keywords: alpha diversity; phylogenetic diversity; rarefaction; sampling depth.
Figures


Similar articles
-
Rarefaction and Extrapolation: Making Fair Comparison of Abundance-Sensitive Phylogenetic Diversity among Multiple Assemblages.Syst Biol. 2017 Jan 1;66(1):100-111. doi: 10.1093/sysbio/syw073. Syst Biol. 2017. PMID: 28173592
-
Multifaceted diversity-area relationships reveal global hotspots of mammalian species, trait and lineage diversity.Glob Ecol Biogeogr. 2014 Aug 1;23(8):836-847. doi: 10.1111/geb.12158. Glob Ecol Biogeogr. 2014. PMID: 25071413 Free PMC article.
-
Abundance-weighted phylogenetic diversity measures distinguish microbial community states and are robust to sampling depth.PeerJ. 2013 Sep 12;1:e157. doi: 10.7717/peerj.157. eCollection 2013. PeerJ. 2013. PMID: 24058885 Free PMC article.
-
Applications of Monte Carlo Simulation in Modelling of Biochemical Processes.In: Mode CJ, editor. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science [Internet]. Rijeka (HR): InTech; 2011 Feb 28. Chapter 4. In: Mode CJ, editor. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science [Internet]. Rijeka (HR): InTech; 2011 Feb 28. Chapter 4. PMID: 28045483 Free Books & Documents. Review.
-
Meta-Analytic Statistical Inferences for Continuous Measure Outcomes as a Function of Effect Size Metric and Other Assumptions [Internet].Rockville (MD): Agency for Healthcare Research and Quality (US); 2013 Apr. Report No.: 13-EHC075-EF. Rockville (MD): Agency for Healthcare Research and Quality (US); 2013 Apr. Report No.: 13-EHC075-EF. PMID: 23741762 Free Books & Documents. Review.
Cited by
-
Pseudomonas spp. diversity is negatively associated with suppression of the wheat take-all pathogen.Sci Rep. 2016 Aug 23;6:29905. doi: 10.1038/srep29905. Sci Rep. 2016. PMID: 27549739 Free PMC article.
-
Energetic and Environmental Constraints on the Community Structure of Benthic Microbial Mats in Lake Fryxell, Antarctica.FEMS Microbiol Ecol. 2020 Feb 1;96(2):fiz207. doi: 10.1093/femsec/fiz207. FEMS Microbiol Ecol. 2020. PMID: 31905236 Free PMC article.
-
Spatial patterns and conservation of genetic and phylogenetic diversity of wildlife in China.Sci Adv. 2021 Jan 22;7(4):eabd5725. doi: 10.1126/sciadv.abd5725. Print 2021 Jan. Sci Adv. 2021. PMID: 33523945 Free PMC article.
-
Species clustering, climate effects, and introduced species in 5 million city trees across 63 US cities.Elife. 2022 Sep 27;11:e77891. doi: 10.7554/eLife.77891. Elife. 2022. PMID: 36165436 Free PMC article.
-
Change in phylogenetic community structure during succession of traditionally managed tropical rainforest in southwest China.PLoS One. 2013 Jul 31;8(7):e71464. doi: 10.1371/journal.pone.0071464. Print 2013. PLoS One. 2013. PMID: 23936268 Free PMC article.
References
-
- Allen B, Kon M, Bar-Yam Y. A new phylogenetic diversity measure generalizing the Shannon index and its application to phyllostomid bats. The American Naturalist. 2009;174(2):236–243. - PubMed
-
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee R, Beck RMD, Grenyer R, Price S, Vos R, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
