Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast
- PMID: 23832786
- PMCID: PMC3725075
- DOI: 10.1073/pnas.1222534110
Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast
Erratum in
-
Correction for Dvir et al., Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast.Proc Natl Acad Sci U S A. 2019 Jul 2;116(27):13701. doi: 10.1073/pnas.1909441116. Epub 2019 Jun 17. Proc Natl Acad Sci U S A. 2019. PMID: 31209029 Free PMC article. No abstract available.
Abstract
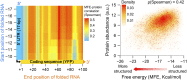
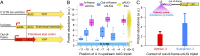
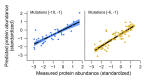
The 5'-untranslated region (5'-UTR) of mRNAs contains elements that affect expression, yet the rules by which these regions exert their effect are poorly understood. Here, we studied the impact of 5'-UTR sequences on protein levels in yeast, by constructing a large-scale library of mutants that differ only in the 10 bp preceding the translational start site of a fluorescent reporter. Using a high-throughput sequencing strategy, we obtained highly accurate measurements of protein abundance for over 2,000 unique sequence variants. The resulting pool spanned an approximately sevenfold range of protein levels, demonstrating the powerful consequences of sequence manipulations of even 1-10 nucleotides immediately upstream of the start codon. We devised computational models that predicted over 70% of the measured expression variability in held-out sequence variants. Notably, a combined model of the most prominent features successfully explained protein abundance in an additional, independently constructed library, whose nucleotide composition differed greatly from the library used to parameterize the model. Our analysis reveals the dominant contribution of the start codon context at positions -3 to -1, mRNA secondary structure, and out-of-frame upstream AUGs (uAUGs) to phenotypic diversity, thereby advancing our understanding of how protein levels are modulated by 5'-UTR sequences, and paving the way toward predictably tuning protein expression through manipulations of 5'-UTRs.
Keywords: AUG sequence context; computational prediction; mRNA folding; post-transcriptional regulation; upstream start codons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Small, synthetic, GC-rich mRNA stem-loop modules 5' proximal to the AUG start-codon predictably tune gene expression in yeast.Microb Cell Fact. 2013 Jul 29;12:74. doi: 10.1186/1475-2859-12-74. Microb Cell Fact. 2013. PMID: 23895661 Free PMC article.
-
Presence of ATG triplets in 5' untranslated regions of eukaryotic cDNAs correlates with a 'weak' context of the start codon.Bioinformatics. 2001 Oct;17(10):890-900. doi: 10.1093/bioinformatics/17.10.890. Bioinformatics. 2001. PMID: 11673233
-
mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae.Mol Microbiol. 1996 Mar;19(6):1225-39. doi: 10.1111/j.1365-2958.1996.tb02468.x. Mol Microbiol. 1996. PMID: 8730865
-
A helicase links upstream ORFs and RNA structure.Curr Genet. 2019 Apr;65(2):453-456. doi: 10.1007/s00294-018-0911-z. Epub 2018 Nov 27. Curr Genet. 2019. PMID: 30483885 Free PMC article. Review.
-
A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3' UTR variants.Hum Genet. 2006 Oct;120(3):301-33. doi: 10.1007/s00439-006-0218-x. Epub 2006 Jun 29. Hum Genet. 2006. PMID: 16807757 Review.
Cited by
-
Functional analysis of the AUG initiator codon context reveals novel conserved sequences that disfavor mRNA translation in eukaryotes.Nucleic Acids Res. 2024 Feb 9;52(3):1064-1079. doi: 10.1093/nar/gkad1152. Nucleic Acids Res. 2024. PMID: 38038264 Free PMC article.
-
Structural imprints in vivo decode RNA regulatory mechanisms.Nature. 2015 Mar 26;519(7544):486-90. doi: 10.1038/nature14263. Epub 2015 Mar 18. Nature. 2015. PMID: 25799993 Free PMC article.
-
Genetic circuit design automation for yeast.Nat Microbiol. 2020 Nov;5(11):1349-1360. doi: 10.1038/s41564-020-0757-2. Epub 2020 Aug 3. Nat Microbiol. 2020. PMID: 32747797
-
Deciphering the rules by which dynamics of mRNA secondary structure affect translation efficiency in Saccharomyces cerevisiae.Nucleic Acids Res. 2014 Apr;42(8):4813-22. doi: 10.1093/nar/gku159. Epub 2014 Feb 21. Nucleic Acids Res. 2014. PMID: 24561808 Free PMC article.
-
Extensive and coordinated control of allele-specific expression by both transcription and translation in Candida albicans.Genome Res. 2014 Jun;24(6):963-73. doi: 10.1101/gr.166322.113. Epub 2014 Apr 14. Genome Res. 2014. PMID: 24732588 Free PMC article.
References
-
- Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44(7):743–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

