Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance
- PMID: 23825228
- PMCID: PMC4046188
- DOI: 10.1096/fj.13-228486
Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance
Abstract
Pathological and physiological stimuli, including acute exercise, activate autophagy; however, it is unknown whether exercise training alters basal levels of autophagy and whether autophagy is required for skeletal muscle adaptation to training. We observed greater autophagy flux (i.e., a combination of increased LC3-II/LC3-I ratio and LC3-II levels and reduced p62 protein content indicating a higher rate of initiation and resolution of autophagic events), autophagy protein expression (i.e., Atg6/Beclin1, Atg7, and Atg8/LC3) and mitophagy protein Bnip3 expression in tonic, oxidative muscle compared to muscles of either mixed fiber types or of predominant glycolytic fibers in mice. Long-term voluntary running (4 wk) resulted in increased basal autophagy flux and expression of autophagy proteins and Bnip3 in parallel to mitochondrial biogenesis in plantaris muscle with mixed fiber types. Conversely, exercise training promoted autophagy protein expression with no significant increases of autophagy flux and mitochondrial biogenesis in the oxidative soleus muscle. We also observed increased basal autophagy flux and Bnip3 content without increases in autophagy protein expression in the plantaris muscle of sedentary muscle-specific Pgc-1α transgenic mice, a genetic model of augmented mitochondrial biogenesis. These findings reveal that endurance exercise training-induced increases in basal autophagy, including mitophagy, only take place if an enhanced oxidative phenotype is achieved. However, autophagy protein expression is mainly dictated by contractile activity independently of enhancements in oxidative phenotype. Exercise-trained mice heterozygous for the critical autophagy protein Atg6 showed attenuated increases of basal autophagy flux, mitochondrial content, and angiogenesis in skeletal muscle, along with impaired improvement of endurance capacity. These results demonstrate that increased basal autophagy is required for endurance exercise training-induced skeletal muscle adaptation and improvement of physical performance.
Keywords: Bnip3; angiogenesis; mitochondrial biogenesis; mitophagy; voluntary wheel running.
Figures
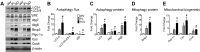
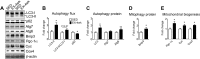
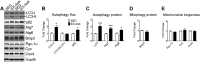
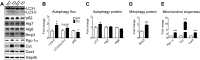

Similar articles
-
Autophagy plays a role in skeletal muscle mitochondrial biogenesis in an endurance exercise-trained condition.J Physiol Sci. 2016 Sep;66(5):417-30. doi: 10.1007/s12576-016-0440-9. Epub 2016 Mar 4. J Physiol Sci. 2016. PMID: 26943341 Free PMC article.
-
Autophagy activation, not peroxisome proliferator-activated receptor γ coactivator 1α, may mediate exercise-induced improvements in glucose handling during diet-induced obesity.Exp Physiol. 2017 Sep 1;102(9):1194-1207. doi: 10.1113/EP086406. Epub 2017 Jul 22. Exp Physiol. 2017. PMID: 28639297
-
Role of Parkin and endurance training on mitochondrial turnover in skeletal muscle.Skelet Muscle. 2018 Mar 17;8(1):10. doi: 10.1186/s13395-018-0157-y. Skelet Muscle. 2018. PMID: 29549884 Free PMC article.
-
The regulation of autophagy during exercise in skeletal muscle.J Appl Physiol (1985). 2016 Mar 15;120(6):664-73. doi: 10.1152/japplphysiol.00550.2015. Epub 2015 Dec 17. J Appl Physiol (1985). 2016. PMID: 26679612 Free PMC article. Review.
-
PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity.Am J Physiol Endocrinol Metab. 2010 Aug;299(2):E145-61. doi: 10.1152/ajpendo.00755.2009. Epub 2010 Apr 6. Am J Physiol Endocrinol Metab. 2010. PMID: 20371735 Free PMC article. Review.
Cited by
-
Protective Effects of Whey Protein Hydrolysate, Treadmill Exercise, and Their Combination against Scopolamine-Induced Cognitive Deficit in Mice.Foods. 2023 Dec 10;12(24):4428. doi: 10.3390/foods12244428. Foods. 2023. PMID: 38137233 Free PMC article.
-
Effects of Exercise Training on the Autophagy-Related Muscular Proteins Expression in Ovariectomized Rats.Front Physiol. 2019 Jun 13;10:735. doi: 10.3389/fphys.2019.00735. eCollection 2019. Front Physiol. 2019. PMID: 31263428 Free PMC article.
-
Autophagy in metabolic disease and ageing.Nat Rev Endocrinol. 2021 Nov;17(11):647-661. doi: 10.1038/s41574-021-00551-9. Epub 2021 Sep 10. Nat Rev Endocrinol. 2021. PMID: 34508250 Review.
-
Mammalian Target of Rapamycin Signaling Pathway Regulates Mitochondrial Quality Control of Brown Adipocytes in Mice.Front Physiol. 2021 Jul 14;12:638352. doi: 10.3389/fphys.2021.638352. eCollection 2021. Front Physiol. 2021. PMID: 34335285 Free PMC article.
-
Effects of aerobic training on markers of autophagy in the elderly.Age (Dordr). 2016 Apr;38(2):33. doi: 10.1007/s11357-016-9897-y. Epub 2016 Mar 3. Age (Dordr). 2016. PMID: 26940016 Free PMC article. Clinical Trial.
References
-
- Manson J. E., Hu F. B., Rich-Edwards J. W., Colditz G. A., Stampfer M. J., Willett W. C., Speizer F. E., Hennekens C. H. (1999) A prospective study of walking as compared with vigorous exercise in the prevention of coronary heart disease in women. N. Engl. J. Med. 341, 650–658 - PubMed
-
- Hu F. B., Manson J. E., Stampfer M. J., Colditz G., Liu S., Solomon C. G., Willett W. C. (2001) Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N. Engl. J. Med. 345, 790–797 - PubMed
-
- Colditz G. A., Cannuscio C. C., Frazier A. L. (1997) Physical activity and reduced risk of colon cancer: implications for prevention. Cancer Causes Control 8, 649–667 - PubMed
-
- Kodama S., Saito K., Tanaka S., Maki M., Yachi Y., Asumi M., Sugawara A., Totsuka K., Shimano H., Ohashi Y., Yamada N., Sone H. (2009) Cardiorespiratory fitness as a quantitative predictor of all-cause mortality and cardiovascular events in healthy men and women: a meta-analysis. JAMA 301, 2024–2035 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

