Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs
- PMID: 23804653
- PMCID: PMC3744946
- DOI: 10.1091/mbc.E13-02-0103
Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs
Abstract
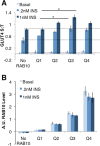
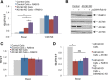
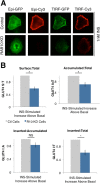
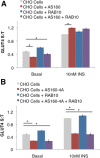
Adipocyte glucose uptake in response to insulin is essential for physiological glucose homeostasis: stimulation of adipocytes with insulin results in insertion of the glucose transporter GLUT4 into the plasma membrane and subsequent glucose uptake. Here we establish that RAB10 and RAB14 are key regulators of GLUT4 trafficking that function at independent, sequential steps of GLUT4 translocation. RAB14 functions upstream of RAB10 in the sorting of GLUT4 to the specialized transport vesicles that ferry GLUT4 to the plasma membrane. RAB10 and its GTPase-activating protein (GAP) AS160 comprise the principal signaling module downstream of insulin receptor activation that regulates the accumulation of GLUT4 transport vesicles at the plasma membrane. Although both RAB10 and RAB14 are regulated by the GAP activity of AS160 in vitro, only RAB10 is under the control of AS160 in vivo. Insulin regulation of the pool of RAB10 required for GLUT4 translocation occurs through regulation of AS160, since activation of RAB10 by DENND4C, its GTP exchange factor, does not require insulin stimulation.
Figures

Similar articles
-
A Rab10:RalA G protein cascade regulates insulin-stimulated glucose uptake in adipocytes.Mol Biol Cell. 2014 Oct 1;25(19):3059-69. doi: 10.1091/mbc.E14-06-1060. Epub 2014 Aug 7. Mol Biol Cell. 2014. PMID: 25103239 Free PMC article.
-
Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane.Cell Metab. 2007 Apr;5(4):293-303. doi: 10.1016/j.cmet.2007.03.001. Cell Metab. 2007. PMID: 17403373
-
Rab10 in insulin-stimulated GLUT4 translocation.Biochem J. 2008 Apr 1;411(1):89-95. doi: 10.1042/BJ20071318. Biochem J. 2008. PMID: 18076383
-
Signal transduction meets vesicle traffic: the software and hardware of GLUT4 translocation.Am J Physiol Cell Physiol. 2014 May 15;306(10):C879-86. doi: 10.1152/ajpcell.00069.2014. Epub 2014 Mar 5. Am J Physiol Cell Physiol. 2014. PMID: 24598362 Review.
-
Glucose transporter 4: cycling, compartments and controversies.EMBO Rep. 2005 Dec;6(12):1137-42. doi: 10.1038/sj.embor.7400584. EMBO Rep. 2005. PMID: 16319959 Free PMC article. Review.
Cited by
-
The Endosomal Protein Endotubin Is Required for Enterocyte Differentiation.Cell Mol Gastroenterol Hepatol. 2017 Nov 15;5(2):145-156. doi: 10.1016/j.jcmgh.2017.11.001. eCollection 2018. Cell Mol Gastroenterol Hepatol. 2017. PMID: 29322087 Free PMC article.
-
Targeting Small GTPases and Their Prenylation in Diabetes Mellitus.J Med Chem. 2021 Jul 22;64(14):9677-9710. doi: 10.1021/acs.jmedchem.1c00410. Epub 2021 Jul 8. J Med Chem. 2021. PMID: 34236862 Free PMC article. Review.
-
Characterization of the Molecular Mechanisms Underlying Glucose Stimulated Insulin Secretion from Isolated Pancreatic β-cells Using Post-translational Modification Specific Proteomics (PTMomics).Mol Cell Proteomics. 2018 Jan;17(1):95-110. doi: 10.1074/mcp.RA117.000217. Epub 2017 Nov 7. Mol Cell Proteomics. 2018. PMID: 29113996 Free PMC article.
-
Chemical biology probes of mammalian GLUT structure and function.Biochem J. 2018 Nov 20;475(22):3511-3534. doi: 10.1042/BCJ20170677. Biochem J. 2018. PMID: 30459202 Free PMC article. Review.
-
Rapamycin Re-Directs Lysosome Network, Stimulates ER-Remodeling, Involving Membrane CD317 and Affecting Exocytosis, in Campylobacter Jejuni-Lysate-Infected U937 Cells.Int J Mol Sci. 2020 Mar 23;21(6):2207. doi: 10.3390/ijms21062207. Int J Mol Sci. 2020. PMID: 32210050 Free PMC article.
References
-
- Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. - PubMed
-
- Bai L, Wang Y, Fan J, Chen Y, Ji W, Qu A, Xu P, James DE, Xu T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metabolism. 2007;5:47–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

