Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains
- PMID: 23791944
- PMCID: PMC3733345
- DOI: 10.1016/j.str.2013.05.008
Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains
Abstract
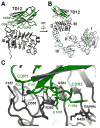
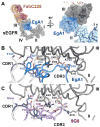
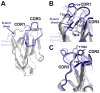
The epidermal growth factor receptor (EGFR) is implicated in human cancers and is the target of several classes of therapeutic agents, including antibody-based drugs. Here, we describe X-ray crystal structures of the extracellular region of EGFR in complex with three inhibitory nanobodies, the variable domains of heavy chain only antibodies (VHH). VHH domains, the smallest natural antigen-binding modules, are readily engineered for diagnostic and therapeutic applications. All three VHH domains prevent ligand-induced EGFR activation, but use two distinct mechanisms. 7D12 sterically blocks ligand binding to EGFR in a manner similar to that of cetuximab. EgA1 and 9G8 bind an epitope near the EGFR domain II/III junction, preventing receptor conformational changes required for high-affinity ligand binding and dimerization. This epitope is accessible to the convex VHH paratope but inaccessible to the flatter paratope of monoclonal antibodies. Appreciating the modes of binding and inhibition of these VHH domains will aid in developing them for tumor imaging and/or cancer therapy.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

Comment in
-
Magic bullets from llamas.Structure. 2013 Jul 2;21(7):1072-3. doi: 10.1016/j.str.2013.06.008. Structure. 2013. PMID: 23823325 Free PMC article.
Similar articles
-
Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization.Cancer Cell. 2008 Apr;13(4):365-73. doi: 10.1016/j.ccr.2008.02.019. Cancer Cell. 2008. PMID: 18394559 Free PMC article.
-
Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.Structure. 2008 Feb;16(2):216-27. doi: 10.1016/j.str.2007.11.009. Structure. 2008. PMID: 18275813
-
A molecular view of anti-ErbB monoclonal antibody therapy.Cancer Cell. 2008 Apr;13(4):291-3. doi: 10.1016/j.ccr.2008.03.010. Cancer Cell. 2008. PMID: 18394550
-
Cetuximab: an epidermal growth factor receptor chemeric human-murine monoclonal antibody.Drugs Today (Barc). 2005 Feb;41(2):107-27. doi: 10.1358/dot.2005.41.2.882662. Drugs Today (Barc). 2005. PMID: 15821783 Review.
-
IRDye 800CW-Anti-epidermal growth factor receptor nanobody 7D12.2012 Apr 6 [updated 2012 Jul 12]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2012 Apr 6 [updated 2012 Jul 12]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 22787693 Free Books & Documents. Review.
Cited by
-
Paper Title "Hu7CG2: A Novel Humanized Anti-Epidermal Growth Factor Receptor (EGFR) Biparatopic Nanobody".Mol Biotechnol. 2021 Jun;63(6):525-533. doi: 10.1007/s12033-021-00317-8. Epub 2021 Mar 26. Mol Biotechnol. 2021. PMID: 33772436
-
Site-Specific Encoding of Photoactivity in Antibodies Enables Light-Mediated Antibody-Antigen Binding on Live Cells.Angew Chem Int Ed Engl. 2019 Dec 9;58(50):17986-17993. doi: 10.1002/anie.201908655. Epub 2019 Oct 31. Angew Chem Int Ed Engl. 2019. PMID: 31609054 Free PMC article.
-
Trispecific T-cell engagers for dual tumor-targeting of colorectal cancer.Oncoimmunology. 2022 Feb 7;11(1):2034355. doi: 10.1080/2162402X.2022.2034355. eCollection 2022. Oncoimmunology. 2022. PMID: 35154908 Free PMC article.
-
Single domain antibodies from camelids in the treatment of microbial infections.Front Immunol. 2024 May 17;15:1334829. doi: 10.3389/fimmu.2024.1334829. eCollection 2024. Front Immunol. 2024. PMID: 38827746 Free PMC article. Review.
-
Nanobody-Based Delivery Systems for Diagnosis and Targeted Tumor Therapy.Front Immunol. 2017 Nov 2;8:1442. doi: 10.3389/fimmu.2017.01442. eCollection 2017. Front Immunol. 2017. PMID: 29163515 Free PMC article. Review.
References
-
- Abbott J, Beckett D. Cooperative binding of the Escherichia coli repressor of biotin biosynthesis to the biotin operator sequence. Biochemistry. 1993;32:9649–9656. - PubMed
-
- Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–2459. - PubMed
-
- Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–352. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

