Whole-genome sequence-based analysis of high-density lipoprotein cholesterol
- PMID: 23770607
- PMCID: PMC4030301
- DOI: 10.1038/ng.2671
Whole-genome sequence-based analysis of high-density lipoprotein cholesterol
Abstract
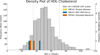
We describe initial steps for interrogating whole-genome sequence data to characterize the genetic architecture of a complex trait, levels of high-density lipoprotein cholesterol (HDL-C). We report whole-genome sequencing and analysis of 962 individuals from the Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) studies. From this analysis, we estimate that common variation contributes more to heritability of HDL-C levels than rare variation, and screening for mendelian variants for dyslipidemia identified individuals with extreme HDL-C levels. Whole-genome sequencing analyses highlight the value of regulatory and non-protein-coding regions of the genome in addition to protein-coding regions.
Figures


Comment in
-
Complex traits: Sequencing for disease architecture.Nat Rev Genet. 2013 Aug;14(8):518. doi: 10.1038/nrg3533. Epub 2013 Jul 2. Nat Rev Genet. 2013. PMID: 23817311 No abstract available.
Similar articles
-
Multiplexed Targeted Resequencing Identifies Coding and Regulatory Variation Underlying Phenotypic Extremes of High-Density Lipoprotein Cholesterol in Humans.Circ Genom Precis Med. 2018 Jul;11(7):e002070. doi: 10.1161/CIRCGEN.117.002070. Circ Genom Precis Med. 2018. PMID: 29987113 Free PMC article.
-
Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels.Arterioscler Thromb Vasc Biol. 2013 Jul;33(7):1521-8. doi: 10.1161/ATVBAHA.113.301505. Epub 2013 May 16. Arterioscler Thromb Vasc Biol. 2013. PMID: 23685560
-
Practical Approaches for Whole-Genome Sequence Analysis of Heart- and Blood-Related Traits.Am J Hum Genet. 2017 Feb 2;100(2):205-215. doi: 10.1016/j.ajhg.2016.12.009. Epub 2017 Jan 12. Am J Hum Genet. 2017. PMID: 28089252 Free PMC article.
-
Genetic-epidemiological evidence on genes associated with HDL cholesterol levels: a systematic in-depth review.Exp Gerontol. 2009 Mar;44(3):136-60. doi: 10.1016/j.exger.2008.11.003. Epub 2008 Nov 17. Exp Gerontol. 2009. PMID: 19041386 Free PMC article. Review.
-
Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population.Atherosclerosis. 2010 Feb;208(2):305-16. doi: 10.1016/j.atherosclerosis.2009.06.005. Epub 2009 Jun 11. Atherosclerosis. 2010. PMID: 19596329 Review.
Cited by
-
Type IV Collagen Variants in CKD: Performance of Computational Predictions for Identifying Pathogenic Variants.Kidney Med. 2021 Feb 10;3(2):257-266. doi: 10.1016/j.xkme.2020.12.007. eCollection 2021 Mar-Apr. Kidney Med. 2021. PMID: 33851121 Free PMC article.
-
Genetic insights into cardiometabolic risk factors.Clin Biochem Rev. 2014 Feb;35(1):15-36. Clin Biochem Rev. 2014. PMID: 24659834 Free PMC article. Review.
-
Rare-variant association analysis: study designs and statistical tests.Am J Hum Genet. 2014 Jul 3;95(1):5-23. doi: 10.1016/j.ajhg.2014.06.009. Am J Hum Genet. 2014. PMID: 24995866 Free PMC article. Review.
-
Association of the IGF1 gene with fasting insulin levels.Eur J Hum Genet. 2016 Aug;24(9):1337-43. doi: 10.1038/ejhg.2016.4. Epub 2016 Feb 10. Eur J Hum Genet. 2016. PMID: 26860063 Free PMC article.
-
Predicting deleterious missense genetic variants via integrative supervised nonnegative matrix tri-factorization.Sci Rep. 2021 Dec 9;11(1):23747. doi: 10.1038/s41598-021-03230-x. Sci Rep. 2021. PMID: 34887492 Free PMC article.
References
-
- Castelli WP, et al. HDL cholesterol and other lipids in coronary heart disease. The cooperative lipoprotein phenotyping study. Circulation. 1977;55:767–772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- N01-HC-85082/HC/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- R01 HL080295/HL/NHLBI NIH HHS/United States
- HL080295/HL/NHLBI NIH HHS/United States
- N01-HC-85084/HC/NHLBI NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- N01-HC-25195/HC/NHLBI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- N01-HC-85085/HC/NHLBI NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- N01-HC-85081/HC/NHLBI NIH HHS/United States
- RC2HL102419-02/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- HL105756/HL/NHLBI NIH HHS/United States
- N01 HC015103/HC/NHLBI NIH HHS/United States
- R56 AG020098/AG/NIA NIH HHS/United States
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- R01 HL072810/HL/NHLBI NIH HHS/United States
- AG-20098/AG/NIA NIH HHS/United States
- HL087652/HL/NHLBI NIH HHS/United States
- N01HC55222/HL/NHLBI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- N01-HC-85086/HC/NHLBI NIH HHS/United States
- N01HC85086/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- AG-027058/AG/NIA NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- N01 HC-55222/HC/NHLBI NIH HHS/United States
- N01-HC-85083/HC/NHLBI NIH HHS/United States
- N01-HC-75150/HC/NHLBI NIH HHS/United States
- N01-HC-85080/HC/NHLBI NIH HHS/United States
- R01 AG020098/AG/NIA NIH HHS/United States
- N01HC75150/HL/NHLBI NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- N01HC25195/HL/NHLBI NIH HHS/United States
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- N01-HC-85079/HC/NHLBI NIH HHS/United States
- N01-HC-85239/HC/NHLBI NIH HHS/United States
- AG-023629/AG/NIA NIH HHS/United States
- N01HC85079/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 AG027058/AG/NIA NIH HHS/United States
- N01 HC045133/HC/NHLBI NIH HHS/United States
- N01 HC035129/HC/NHLBI NIH HHS/United States
- R56 AG023629/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

