Direct transcriptional reprogramming of adult cells to embryonic nephron progenitors
- PMID: 23766537
- PMCID: PMC3752949
- DOI: 10.1681/ASN.2012121143
Direct transcriptional reprogramming of adult cells to embryonic nephron progenitors
Abstract
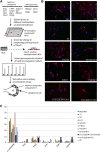
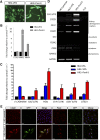
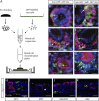
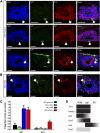
Direct reprogramming involves the enforced re-expression of key transcription factors to redefine a cellular state. The nephron progenitor population of the embryonic kidney gives rise to all cells within the nephron other than the collecting duct through a mesenchyme-to-epithelial transition, but this population is exhausted around the time of birth. Here, we sought to identify the conditions under which adult proximal tubule cells could be directly transcriptionally reprogrammed to nephron progenitors. Using a combinatorial screen for lineage-instructive transcription factors, we identified a pool of six genes (SIX1, SIX2, OSR1, EYA1, HOXA11, and SNAI2) that activated a network of genes consistent with a cap mesenchyme/nephron progenitor phenotype in the adult proximal tubule (HK2) cell line. Consistent with these reprogrammed cells being nephron progenitors, we observed differential contribution of the reprogrammed population into the Six2(+) nephron progenitor fields of an embryonic kidney explant. Dereplication of the pool suggested that SNAI2 can suppress E-CADHERIN, presumably assisting in the epithelial-to-mesenchymal transition (EMT) required to form nephron progenitors. However, neither TGFβ-induced EMT nor SNAI2 overexpression alone was sufficient to create this phenotype, suggesting that additional factors are required. In conclusion, these results suggest that reinitiation of kidney development from a population of adult cells by generating embryonic progenitors may be feasible, opening the way for additional cellular and bioengineering approaches to renal repair and regeneration.
Figures

Similar articles
-
Osr1 acts downstream of and interacts synergistically with Six2 to maintain nephron progenitor cells during kidney organogenesis.Development. 2014 Apr;141(7):1442-52. doi: 10.1242/dev.103283. Epub 2014 Mar 5. Development. 2014. PMID: 24598167 Free PMC article.
-
Direct reprogramming to human nephron progenitor-like cells using inducible piggyBac transposon expression of SNAI2-EYA1-SIX1.Kidney Int. 2019 May;95(5):1153-1166. doi: 10.1016/j.kint.2018.11.041. Epub 2019 Feb 28. Kidney Int. 2019. PMID: 30827514 Free PMC article.
-
Chromatin Remodelers Interact with Eya1 and Six2 to Target Enhancers to Control Nephron Progenitor Cell Maintenance.J Am Soc Nephrol. 2021 Nov;32(11):2815-2833. doi: 10.1681/ASN.2021040525. J Am Soc Nephrol. 2021. PMID: 34716243 Free PMC article.
-
Nephron progenitor cells: shifting the balance of self-renewal and differentiation.Curr Top Dev Biol. 2014;107:293-331. doi: 10.1016/B978-0-12-416022-4.00011-1. Curr Top Dev Biol. 2014. PMID: 24439811 Review.
-
Epigenetic States of nephron progenitors and epithelial differentiation.J Cell Biochem. 2015 Jun;116(6):893-902. doi: 10.1002/jcb.25048. J Cell Biochem. 2015. PMID: 25560433 Free PMC article. Review.
Cited by
-
Induction of human pluripotent stem cells into kidney tissues by synthetic mRNAs encoding transcription factors.Sci Rep. 2019 Jan 29;9(1):913. doi: 10.1038/s41598-018-37485-8. Sci Rep. 2019. PMID: 30696889 Free PMC article.
-
Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks.Development. 2016 Aug 1;143(15):2696-705. doi: 10.1242/dev.138263. Development. 2016. PMID: 27486230 Free PMC article. Review.
-
Growing a new human kidney.Kidney Int. 2019 Oct;96(4):871-882. doi: 10.1016/j.kint.2019.04.040. Epub 2019 May 25. Kidney Int. 2019. PMID: 31399199 Free PMC article. Review.
-
Direct reprogramming of human bone marrow stromal cells into functional renal cells using cell-free extracts.Stem Cell Reports. 2015 Apr 14;4(4):685-98. doi: 10.1016/j.stemcr.2015.02.002. Epub 2015 Mar 5. Stem Cell Reports. 2015. PMID: 25754206 Free PMC article.
-
Wound healing applications of creams and "smart" hydrogels.Exp Dermatol. 2021 Sep;30(9):1218-1232. doi: 10.1111/exd.14396. Epub 2021 Jun 7. Exp Dermatol. 2021. PMID: 34009648 Free PMC article. Review.
References
-
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L: Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475: 386–389, 2011 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

