Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling
- PMID: 23763768
- PMCID: PMC3850062
- DOI: 10.1016/j.virol.2013.04.019
Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling
Abstract
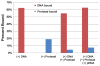
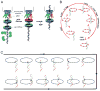
It has long been believed that the DNA-packaging motor of dsDNA viruses utilizes a rotation mechanism. Here we report a revolution rather than rotation mechanism for the bacteriophage phi29 DNA packaging motor. The phi29 motor contains six copies of the ATPase (Schwartz et al., this issue); ATP binding to one ATPase subunit stimulates the ATPase to adopt a conformation with a high affinity for dsDNA. ATP hydrolysis induces a new conformation with a lower affinity, thus transferring the dsDNA to an adjacent subunit by a power stroke. DNA revolves unidirectionally along the hexameric channel wall of the ATPase, but neither the dsDNA nor the ATPase itself rotates along its own axis. One ATP is hydrolyzed in each transitional step, and six ATPs are consumed for one helical turn of 360°. Transition of the same dsDNA chain along the channel wall, but at a location 60° different from the last contact, urges dsDNA to move forward 1.75 base pairs each step (10.5bp per turn/6ATP=1.75bp per ATP). Each connector subunit tilts with a left-handed orientation at a 30° angle in relation to its vertical axis that runs anti-parallel to the right-handed dsDNA helix, facilitating the one-way traffic of dsDNA. The connector channel has been shown to cause four steps of transition due to four positively charged lysine rings that make direct contact with the negatively charged DNA phosphate backbone. Translocation of dsDNA into the procapsid by revolution avoids the difficulties during rotation that are associated with DNA supercoiling. Since the revolution mechanism can apply to any stoichiometry, this motor mechanism might reconcile the stoichiometry discrepancy in many phage systems where the ATPase has been found as a tetramer, hexamer, or nonamer.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Ultrastable pRNA hexameric ring gearing hexameric phi29 DNA-packaging motor by revolving without rotating and coiling.Curr Opin Biotechnol. 2013 Aug;24(4):581-90. doi: 10.1016/j.copbio.2013.03.019. Epub 2013 May 14. Curr Opin Biotechnol. 2013. PMID: 23683853 Free PMC article. Review.
-
The ATPase of the phi29 DNA packaging motor is a member of the hexameric AAA+ superfamily.Virology. 2013 Aug 15;443(1):20-7. doi: 10.1016/j.virol.2013.04.004. Epub 2013 May 22. Virology. 2013. PMID: 23706809 Free PMC article.
-
"Push through one-way valve" mechanism of viral DNA packaging.Adv Virus Res. 2012;83:415-65. doi: 10.1016/B978-0-12-394438-2.00009-8. Adv Virus Res. 2012. PMID: 22748815 Review.
-
Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging.Nucleic Acids Res. 2012 Mar;40(6):2577-86. doi: 10.1093/nar/gkr841. Epub 2011 Nov 22. Nucleic Acids Res. 2012. PMID: 22110031 Free PMC article.
-
Mechanism of one-way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating antiparallel revolution.ACS Nano. 2013 May 28;7(5):4082-92. doi: 10.1021/nn4002775. Epub 2013 Mar 26. ACS Nano. 2013. PMID: 23510192 Free PMC article.
Cited by
-
Elastic properties and heterogeneous stiffness of the phi29 motor connector channel.Biophys J. 2014 Mar 18;106(6):1338-48. doi: 10.1016/j.bpj.2014.01.028. Biophys J. 2014. PMID: 24655509 Free PMC article.
-
Progress towards revealing the mechanism of herpesvirus capsid maturation and genome packaging.Protein Cell. 2020 May;11(5):316-317. doi: 10.1007/s13238-020-00716-8. Protein Cell. 2020. PMID: 32270449 Free PMC article. No abstract available.
-
Packaging Models versus Modeling Packaging.Biophys J. 2016 Jan 19;110(2):287-288. doi: 10.1016/j.bpj.2015.10.056. Biophys J. 2016. PMID: 26789751 Free PMC article. No abstract available.
-
Biological Nanomotors with a Revolution, Linear, or Rotation Motion Mechanism.Microbiol Mol Biol Rev. 2016 Jan 27;80(1):161-86. doi: 10.1128/MMBR.00056-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26819321 Free PMC article. Review.
-
Ultrastable pRNA hexameric ring gearing hexameric phi29 DNA-packaging motor by revolving without rotating and coiling.Curr Opin Biotechnol. 2013 Aug;24(4):581-90. doi: 10.1016/j.copbio.2013.03.019. Epub 2013 May 14. Curr Opin Biotechnol. 2013. PMID: 23683853 Free PMC article. Review.
References
-
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. - PubMed
-
- Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. - PubMed
-
- Badasso MO, Leiman PG, Tao Y, He Y, Ohlendorf DH, Rossmann MG, Anderson D. Purification, crystallization and initial X-ray analysis of the head- tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2000;56 (Pt 9):1187–1190. - PubMed
-
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

