Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer
- PMID: 23757493
- PMCID: PMC3725050
- DOI: 10.1073/pnas.1307382110
Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer
Abstract
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer, a membrane-fusing machine, mediates virus entry into host cells and is the sole virus-specific target for neutralizing antibodies. Binding the receptors, CD4 and CCR5/CXCR4, triggers Env conformational changes from the metastable unliganded state to the fusion-active state. We used cryo-electron microscopy to obtain a 6-Å structure of the membrane-bound, heavily glycosylated HIV-1 Env trimer in its uncleaved and unliganded state. The spatial organization of secondary structure elements reveals that the unliganded conformations of both glycoprotein (gp)120 and gp41 subunits differ from those induced by receptor binding. The gp120 trimer association domains, which contribute to interprotomer contacts in the unliganded Env trimer, undergo rearrangement upon CD4 binding. In the unliganded Env, intersubunit interactions maintain the gp41 ectodomain helical bundles in a "spring-loaded" conformation distinct from the extended helical coils of the fusion-active state. Quaternary structure regulates the virus-neutralizing potency of antibodies targeting the conserved CD4-binding site on gp120. The Env trimer architecture provides mechanistic insights into the metastability of the unliganded state, receptor-induced conformational changes, and quaternary structure-based strategies for immune evasion.
Keywords: cryo-EM; membrane protein; retrovirus; spike; vaccine immunogen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
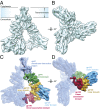
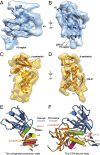
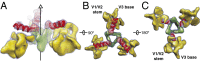

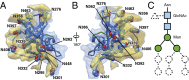
Comment in
-
Finding trimeric HIV-1 envelope glycoproteins in random noise.Proc Natl Acad Sci U S A. 2013 Nov 5;110(45):E4175-7. doi: 10.1073/pnas.1314353110. Epub 2013 Oct 8. Proc Natl Acad Sci U S A. 2013. PMID: 24106301 Free PMC article. No abstract available.
-
Structure of trimeric HIV-1 envelope glycoproteins.Proc Natl Acad Sci U S A. 2013 Nov 5;110(45):E4172-4. doi: 10.1073/pnas.1313802110. Epub 2013 Oct 8. Proc Natl Acad Sci U S A. 2013. PMID: 24106302 Free PMC article. No abstract available.
-
Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise.Proc Natl Acad Sci U S A. 2013 Nov 5;110(45):18037-41. doi: 10.1073/pnas.1314449110. Epub 2013 Oct 8. Proc Natl Acad Sci U S A. 2013. PMID: 24106306 Free PMC article.
-
Reply to Subramaniam, van Heel, and Henderson: Validity of the cryo-electron microscopy structures of the HIV-1 envelope glycoprotein complex.Proc Natl Acad Sci U S A. 2013 Nov 5;110(45):E4178-82. doi: 10.1073/pnas.1316666110. Proc Natl Acad Sci U S A. 2013. PMID: 24350339 Free PMC article. No abstract available.
Similar articles
-
Residues in the gp41 Ectodomain Regulate HIV-1 Envelope Glycoprotein Conformational Transitions Induced by gp120-Directed Inhibitors.J Virol. 2017 Feb 14;91(5):e02219-16. doi: 10.1128/JVI.02219-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28003492 Free PMC article.
-
Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer.PLoS One. 2015 Apr 7;10(4):e0122111. doi: 10.1371/journal.pone.0122111. eCollection 2015. PLoS One. 2015. PMID: 25849367 Free PMC article.
-
Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):E7151-E7158. doi: 10.1073/pnas.1615939113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799557 Free PMC article.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
-
HIV-1 envelope glycoprotein structure.Curr Opin Struct Biol. 2013 Apr;23(2):268-76. doi: 10.1016/j.sbi.2013.03.007. Epub 2013 Apr 18. Curr Opin Struct Biol. 2013. PMID: 23602427 Free PMC article. Review.
Cited by
-
Ferritin-based nanomedicine for disease treatment.Med Rev (2021). 2023 Mar 10;3(1):49-74. doi: 10.1515/mr-2023-0001. eCollection 2023 Feb. Med Rev (2021). 2023. PMID: 37724111 Free PMC article. Review.
-
Improvement of cryo-EM maps by simultaneous local and non-local deep learning.Nat Commun. 2023 Jun 3;14(1):3217. doi: 10.1038/s41467-023-39031-1. Nat Commun. 2023. PMID: 37270635 Free PMC article.
-
EMPIAR: the Electron Microscopy Public Image Archive.Nucleic Acids Res. 2023 Jan 6;51(D1):D1503-D1511. doi: 10.1093/nar/gkac1062. Nucleic Acids Res. 2023. PMID: 36440762 Free PMC article.
-
Membrane attachment and fusion of HIV-1, influenza A, and SARS-CoV-2: resolving the mechanisms with biophysical methods.Biophys Rev. 2022 Oct 11;14(5):1109-1140. doi: 10.1007/s12551-022-00999-7. eCollection 2022 Oct. Biophys Rev. 2022. PMID: 36249860 Free PMC article. Review.
-
Residues L55 and W69 of Tva Mediate Entry of Subgroup A Avian Leukosis Virus.J Virol. 2022 Sep 28;96(18):e0067822. doi: 10.1128/jvi.00678-22. Epub 2022 Sep 7. J Virol. 2022. PMID: 36069550 Free PMC article.
References
-
- Barré-Sinoussi F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. - PubMed
-
- Gallo RC, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224(4648):500–503. - PubMed
-
- Allan JS, et al. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228(4703):1091–1094. - PubMed
-
- Robey WG, et al. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science. 1985;228(4699):593–595. - PubMed
-
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

