Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism
- PMID: 23754953
- PMCID: PMC3675007
- DOI: 10.1371/journal.pgen.1003523
Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism
Abstract
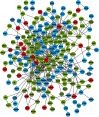
Autism Spectrum Disorders (ASD) are highly heritable and characterised by impairments in social interaction and communication, and restricted and repetitive behaviours. Considering four sets of de novo copy number variants (CNVs) identified in 181 individuals with autism and exploiting mouse functional genomics and known protein-protein interactions, we identified a large and significantly interconnected interaction network. This network contains 187 genes affected by CNVs drawn from 45% of the patients we considered and 22 genes previously implicated in ASD, of which 192 form a single interconnected cluster. On average, those patients with copy number changed genes from this network possess changes in 3 network genes, suggesting that epistasis mediated through the network is extensive. Correspondingly, genes that are highly connected within the network, and thus whose copy number change is predicted by the network to be more phenotypically consequential, are significantly enriched among patients that possess only a single ASD-associated network copy number changed gene (p = 0.002). Strikingly, deleted or disrupted genes from the network are significantly enriched in GO-annotated positive regulators (2.3-fold enrichment, corrected p = 2×10(-5)), whereas duplicated genes are significantly enriched in GO-annotated negative regulators (2.2-fold enrichment, corrected p = 0.005). The direction of copy change is highly informative in the context of the network, providing the means through which perturbations arising from distinct deletions or duplications can yield a common outcome. These findings reveal an extensive ASD-associated molecular network, whose topology indicates ASD-relevant mutational deleteriousness and that mechanistically details how convergent aetiologies can result extensively from CNVs affecting pathways causally implicated in ASD.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Synergistic interactions between Drosophila orthologues of genes spanned by de novo human CNVs support multiple-hit models of autism.PLoS Genet. 2015 Mar 27;11(3):e1004998. doi: 10.1371/journal.pgen.1004998. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25816101 Free PMC article.
-
Functional impact of global rare copy number variation in autism spectrum disorders.Nature. 2010 Jul 15;466(7304):368-72. doi: 10.1038/nature09146. Epub 2010 Jun 9. Nature. 2010. PMID: 20531469 Free PMC article.
-
Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts.Mol Psychiatry. 2013 Oct;18(10):1090-5. doi: 10.1038/mp.2012.138. Epub 2012 Oct 9. Mol Psychiatry. 2013. PMID: 23044707 Free PMC article.
-
Genetic architecture in autism spectrum disorder.Curr Opin Genet Dev. 2012 Jun;22(3):229-37. doi: 10.1016/j.gde.2012.03.002. Epub 2012 Mar 29. Curr Opin Genet Dev. 2012. PMID: 22463983 Review.
-
[Autism spectrum disorder and genes for synaptic proteins].Brain Nerve. 2012 Jan;64(1):65-70. Brain Nerve. 2012. PMID: 22223503 Review. Japanese.
Cited by
-
Duplications in ADHD patients harbour neurobehavioural genes that are co-expressed with genes associated with hyperactivity in the mouse.Am J Med Genet B Neuropsychiatr Genet. 2015 Mar;168B(2):97-107. doi: 10.1002/ajmg.b.32285. Epub 2015 Feb 5. Am J Med Genet B Neuropsychiatr Genet. 2015. PMID: 25656289 Free PMC article.
-
Gene networks underlying convergent and pleiotropic phenotypes in a large and systematically-phenotyped cohort with heterogeneous developmental disorders.PLoS Genet. 2015 Mar 17;11(3):e1005012. doi: 10.1371/journal.pgen.1005012. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25781962 Free PMC article.
-
Integrated systems analysis reveals a molecular network underlying autism spectrum disorders.Mol Syst Biol. 2014 Dec 30;10(12):774. doi: 10.15252/msb.20145487. Mol Syst Biol. 2014. PMID: 25549968 Free PMC article.
-
Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism.Nat Commun. 2014 Apr 11;5:3650. doi: 10.1038/ncomms4650. Nat Commun. 2014. PMID: 24722188 Free PMC article.
-
Identification of Human Neuronal Protein Complexes Reveals Biochemical Activities and Convergent Mechanisms of Action in Autism Spectrum Disorders.Cell Syst. 2015 Nov 25;1(5):361-374. doi: 10.1016/j.cels.2015.11.002. Cell Syst. 2015. PMID: 26949739 Free PMC article.
References
-
- Kogan MD, Blumberg SJ, Schieve LA, Boyle CA, Perrin JM, et al. (2009) Prevalence of parent-reported diagnosis of autism spectrum disorder among children in the US, 2007. Pediatrics 124: 1395–1403. - PubMed
-
- Veenstra-Vanderweele J, Christian SL, Cook EH Jr (2004) Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet 5: 379–405. - PubMed
-
- Chakrabarti S, Fombonne E (2005) Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry 162: 1133–1141. - PubMed
-
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25: 63–77. - PubMed
-
- Stankiewicz P, Lupski JR (2010) Structural variation in the human genome and its role in disease. Annu Rev Med 61: 437–455. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

