RBPmotif: a web server for the discovery of sequence and structure preferences of RNA-binding proteins
- PMID: 23754853
- PMCID: PMC3692078
- DOI: 10.1093/nar/gkt463
RBPmotif: a web server for the discovery of sequence and structure preferences of RNA-binding proteins
Abstract
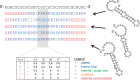
RBPmotif web server (http://www.rnamotif.org) implements tools to identify binding preferences of RNA-binding proteins (RBPs). Given a set of sequences that are known to be bound and unbound by the RBP of interest, RBPmotif provides two types of analysis: (i) de novo motif finding when there is no a priori knowledge on RBP's binding preferences and (ii) analysis of structure preferences when there is a previously identified sequence motif for the RBP. De novo motif finding is performed with the previously published RNAcontext algorithm that learns discriminative motif models to identify both sequence and structure preferences. The results of this analysis include the inferred binding preferences of the RBP and the added predictive value of incorporating structure preferences. Second type of analysis investigates whether the instances of the previously identified sequence motif are enriched in a particular structure context in bound sequences, relative to its instances in unbound sequences. On completion, the results page shows the comparison of structure contexts of the motif instances between bound and unbound sequences and an assessment of statistical significance of detected preferences. In summary, RBPmotif web server enables the concurrent analysis of sequence and structure preferences of RBPs through a user-friendly interface.
Figures

Similar articles
-
Finding the target sites of RNA-binding proteins.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):111-30. doi: 10.1002/wrna.1201. Epub 2013 Nov 11. Wiley Interdiscip Rev RNA. 2014. PMID: 24217996 Free PMC article. Review.
-
RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins.PLoS Comput Biol. 2010 Jul 1;6(7):e1000832. doi: 10.1371/journal.pcbi.1000832. PLoS Comput Biol. 2010. PMID: 20617199 Free PMC article.
-
BRIO: a web server for RNA sequence and structure motif scan.Nucleic Acids Res. 2021 Jul 2;49(W1):W67-W71. doi: 10.1093/nar/gkab400. Nucleic Acids Res. 2021. PMID: 34038531 Free PMC article.
-
SMARTIV: combined sequence and structure de-novo motif discovery for in-vivo RNA binding data.Nucleic Acids Res. 2018 Jul 2;46(W1):W221-W228. doi: 10.1093/nar/gky453. Nucleic Acids Res. 2018. PMID: 29800452 Free PMC article.
-
Motif models for RNA-binding proteins.Curr Opin Struct Biol. 2018 Dec;53:115-123. doi: 10.1016/j.sbi.2018.08.001. Epub 2018 Aug 29. Curr Opin Struct Biol. 2018. PMID: 30172081 Review.
Cited by
-
Computational Methods for CLIP-seq Data Processing.Bioinform Biol Insights. 2014 Oct 1;8:199-207. doi: 10.4137/BBI.S16803. eCollection 2014. Bioinform Biol Insights. 2014. PMID: 25336930 Free PMC article. Review.
-
RNA Thermodynamic Structural Entropy.PLoS One. 2015 Nov 10;10(11):e0137859. doi: 10.1371/journal.pone.0137859. eCollection 2015. PLoS One. 2015. PMID: 26555444 Free PMC article.
-
GraphProt: modeling binding preferences of RNA-binding proteins.Genome Biol. 2014 Jan 22;15(1):R17. doi: 10.1186/gb-2014-15-1-r17. Genome Biol. 2014. PMID: 24451197 Free PMC article.
-
Finding the target sites of RNA-binding proteins.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):111-30. doi: 10.1002/wrna.1201. Epub 2013 Nov 11. Wiley Interdiscip Rev RNA. 2014. PMID: 24217996 Free PMC article. Review.
-
High-throughput characterization of protein-RNA interactions.Brief Funct Genomics. 2015 Jan;14(1):74-89. doi: 10.1093/bfgp/elu047. Epub 2014 Dec 13. Brief Funct Genomics. 2015. PMID: 25504152 Free PMC article. Review.
References
-
- Ray D, Kazan H, Chan E, Castillo L, Chaudhry S, Talukder S, Blencowe B, Morris Q, Hughes T. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. - PubMed
-
- Ule J, Jensen K, Mele A, Darnell R. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. - PubMed
-
- Aviv T, Lin Z, Ben-Ari G, Smibert C, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1. Nat. Struct. Mol. Biol. 2006;13:168–176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

