OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin
- PMID: 23746843
- PMCID: PMC3690481
- DOI: 10.1016/j.cell.2013.05.014
OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin
Abstract
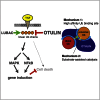
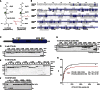
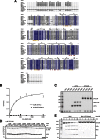
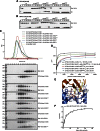
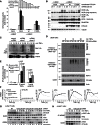
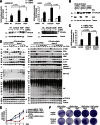
The linear ubiquitin (Ub) chain assembly complex (LUBAC) is an E3 ligase that specifically assembles Met1-linked (also known as linear) Ub chains that regulate nuclear factor κB (NF-κB) signaling. Deubiquitinases (DUBs) are key regulators of Ub signaling, but a dedicated DUB for Met1 linkages has not been identified. Here, we reveal a previously unannotated human DUB, OTULIN (also known as FAM105B), which is exquisitely specific for Met1 linkages. Crystal structures of the OTULIN catalytic domain in complex with diubiquitin reveal Met1-specific Ub-binding sites and a mechanism of substrate-assisted catalysis in which the proximal Ub activates the catalytic triad of the protease. Mutation of Ub Glu16 inhibits OTULIN activity by reducing kcat 240-fold. OTULIN overexpression or knockdown affects NF-κB responses to LUBAC, TNFα, and poly(I:C) and sensitizes cells to TNFα-induced cell death. We show that OTULIN binds LUBAC and that overexpression of OTULIN prevents TNFα-induced NEMO association with ubiquitinated RIPK1. Our data suggest that OTULIN regulates Met1-polyUb signaling.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



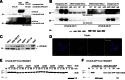
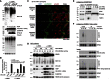
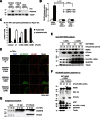

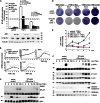
Comment in
-
A new DUB makes linear ubiquitin a party to its own destruction.Cell. 2013 Jun 6;153(6):1189-91. doi: 10.1016/j.cell.2013.05.018. Cell. 2013. PMID: 23746836
-
Post-translational modifications: Breaking linear chains.Nat Rev Mol Cell Biol. 2013 Jul;14(7):402-3. doi: 10.1038/nrm3612. Epub 2013 Jun 19. Nat Rev Mol Cell Biol. 2013. PMID: 23778969 No abstract available.
Similar articles
-
Regulation of Met1-linked polyubiquitin signalling by the deubiquitinase OTULIN.FEBS J. 2016 Jan;283(1):39-53. doi: 10.1111/febs.13547. Epub 2015 Nov 2. FEBS J. 2016. PMID: 26503766 Free PMC article. Review.
-
Molecular basis and regulation of OTULIN-LUBAC interaction.Mol Cell. 2014 May 8;54(3):335-48. doi: 10.1016/j.molcel.2014.03.018. Epub 2014 Apr 10. Mol Cell. 2014. PMID: 24726323 Free PMC article.
-
OTULIN restricts Met1-linked ubiquitination to control innate immune signaling.Mol Cell. 2013 Jun 27;50(6):818-830. doi: 10.1016/j.molcel.2013.06.004. Mol Cell. 2013. PMID: 23806334 Free PMC article.
-
CYLD Limits Lys63- and Met1-Linked Ubiquitin at Receptor Complexes to Regulate Innate Immune Signaling.Cell Rep. 2016 Mar 29;14(12):2846-58. doi: 10.1016/j.celrep.2016.02.062. Epub 2016 Mar 17. Cell Rep. 2016. PMID: 26997266 Free PMC article.
-
Ubiquitin-dependent and -independent functions of OTULIN in cell fate control and beyond.Cell Death Differ. 2021 Feb;28(2):493-504. doi: 10.1038/s41418-020-00675-x. Epub 2020 Dec 7. Cell Death Differ. 2021. PMID: 33288901 Free PMC article. Review.
Cited by
-
In-Plate Chemical Synthesis of Isopeptide-Linked SUMOylated Peptide Fluorescence Polarization Reagents for High-Throughput Screening of SENP Preferences.Chembiochem. 2023 Jan 17;24(2):e202200601. doi: 10.1002/cbic.202200601. Epub 2022 Dec 7. Chembiochem. 2023. PMID: 36377600 Free PMC article.
-
The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases.Mol Cell. 2016 Jul 21;63(2):261-276. doi: 10.1016/j.molcel.2016.06.015. Epub 2016 Jul 14. Mol Cell. 2016. PMID: 27425412 Free PMC article.
-
The centrosomal deubiquitylase USP21 regulates Gli1 transcriptional activity and stability.J Cell Sci. 2016 Nov 1;129(21):4001-4013. doi: 10.1242/jcs.188516. Epub 2016 Sep 12. J Cell Sci. 2016. PMID: 27621083 Free PMC article.
-
Mechanism of Lys6 poly-ubiquitin specificity by the L. pneumophila deubiquitinase LotA.Mol Cell. 2023 Jan 5;83(1):105-120.e5. doi: 10.1016/j.molcel.2022.11.022. Epub 2022 Dec 19. Mol Cell. 2023. PMID: 36538933 Free PMC article.
-
A bio-orthogonal linear ubiquitin probe identifies STAT3 as a direct substrate of OTULIN in glioblastoma.Nucleic Acids Res. 2023 Feb 22;51(3):1050-1066. doi: 10.1093/nar/gkad002. Nucleic Acids Res. 2023. PMID: 36660824 Free PMC article.
References
-
- Behrends C., Harper J.W. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 2011;18:520–528. - PubMed
-
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. - PubMed
-
- Bucher P., Karplus K., Moeri N., Hofmann K. A flexible motif search technique based on generalized profiles. Comput. Chem. 1996;20:3–23. - PubMed
Supplemental References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

