Structural mechanism of cytosolic DNA sensing by cGAS
- PMID: 23722159
- PMCID: PMC3768140
- DOI: 10.1038/nature12305
Structural mechanism of cytosolic DNA sensing by cGAS
Abstract
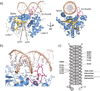
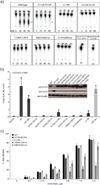
Cytosolic DNA arising from intracellular bacterial or viral infections is a powerful pathogen-associated molecular pattern (PAMP) that leads to innate immune host defence by the production of type I interferon and inflammatory cytokines. Recognition of cytosolic DNA by the recently discovered cyclic-GMP-AMP (cGAMP) synthase (cGAS) induces the production of cGAMP to activate the stimulator of interferon genes (STING). Here we report the crystal structure of cGAS alone and in complex with DNA, ATP and GTP along with functional studies. Our results explain the broad DNA sensing specificity of cGAS, show how cGAS catalyses dinucleotide formation and indicate activation by a DNA-induced structural switch. cGAS possesses a remarkable structural similarity to the antiviral cytosolic double-stranded RNA sensor 2'-5'oligoadenylate synthase (OAS1), but contains a unique zinc thumb that recognizes B-form double-stranded DNA. Our results mechanistically unify dsRNA and dsDNA innate immune sensing by OAS1 and cGAS nucleotidyl transferases.
Figures

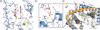
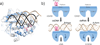
Similar articles
-
Structural and functional analyses of DNA-sensing and immune activation by human cGAS.PLoS One. 2013 Oct 7;8(10):e76983. doi: 10.1371/journal.pone.0076983. eCollection 2013. PLoS One. 2013. PMID: 24116191 Free PMC article.
-
The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop.Cell Rep. 2014 Feb 13;6(3):421-30. doi: 10.1016/j.celrep.2014.01.003. Epub 2014 Jan 23. Cell Rep. 2014. PMID: 24462292 Free PMC article.
-
Unraveling the cGAS catalytic mechanism upon DNA activation through molecular dynamics simulations.Phys Chem Chem Phys. 2021 Apr 22;23(15):9524-9531. doi: 10.1039/d1cp00378j. Phys Chem Chem Phys. 2021. PMID: 33885101
-
The cGAS-STING pathway: The role of self-DNA sensing in inflammatory lung disease.FASEB J. 2020 Oct;34(10):13156-13170. doi: 10.1096/fj.202001607R. Epub 2020 Aug 28. FASEB J. 2020. PMID: 32860267 Free PMC article. Review.
-
Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA.Annu Rev Biochem. 2017 Jun 20;86:541-566. doi: 10.1146/annurev-biochem-061516-044813. Epub 2017 Apr 7. Annu Rev Biochem. 2017. PMID: 28399655 Review.
Cited by
-
Structural basis for sequestration and autoinhibition of cGAS by chromatin.Nature. 2020 Nov;587(7835):678-682. doi: 10.1038/s41586-020-2748-0. Epub 2020 Sep 10. Nature. 2020. PMID: 32911480
-
Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing.Nat Immunol. 2016 Sep 20;17(10):1142-9. doi: 10.1038/ni.3558. Nat Immunol. 2016. PMID: 27648547 Review.
-
PQBP1 Is a Proximal Sensor of the cGAS-Dependent Innate Response to HIV-1.Cell. 2015 Jun 4;161(6):1293-1305. doi: 10.1016/j.cell.2015.04.050. Cell. 2015. PMID: 26046437 Free PMC article.
-
Role of released mitochondrial DNA in acute lung injury.Front Immunol. 2022 Aug 18;13:973089. doi: 10.3389/fimmu.2022.973089. eCollection 2022. Front Immunol. 2022. PMID: 36059472 Free PMC article. Review.
-
Molecular Function of cGAS-STING in SARS-CoV-2: A Novel Approach to COVID-19 Treatment.Biomed Res Int. 2022 Nov 22;2022:6189254. doi: 10.1155/2022/6189254. eCollection 2022. Biomed Res Int. 2022. PMID: 36457340 Free PMC article. Review.
References
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
-
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. - PubMed
-
- Krug A. Nucleic acid recognition receptors in autoimmunity. Handb Exp Pharmacol. 2008:129–151. - PubMed
-
- Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. - PubMed
Methods References
-
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. - PubMed
-
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. - PubMed
-
- Morris RJ, Perrakis A, Lamzin VS. ARP/wARP's model-building algorithms.I. The main chain. Acta Crystallogr D Biol Crystallogr. 2002;58:968–975. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

