Serum response factor indirectly regulates type I interferon-signaling in macrophages
- PMID: 23705899
- PMCID: PMC3793656
- DOI: 10.1089/jir.2012.0065
Serum response factor indirectly regulates type I interferon-signaling in macrophages
Abstract
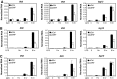
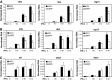
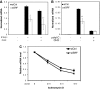
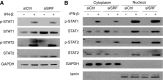
Serum response factor (SRF) is required for diverse aspects of development and homeostasis, but potential roles in the regulation of inflammation and immunity have not been systematically investigated. Here, we demonstrate that SRF is unexpectedly required for optimal responses of elicited peritoneal macrophages to type I interferons. Knockdown of SRF expression in these cells impairs induction of numerous interferon (IFN)-stimulated genes (ISGs) in response to zymosan, LPS, and poly I:C. This effect is primarily due to a defect in the ability of induced type I interferons to mediate secondary activation of ISGs. SRF does not appear to be required for expression of established components of the type I interferon signaling pathway, with IFN-β-dependent phosphorylation of STAT1 and STAT2 normally occurring in SRF-depleted macrophages. Collectively, these findings suggest that SRF can indirectly modulate type I interferon-signaling, without interfering with the classic JAK/STAT/ISGF3 pathway.
Figures

Similar articles
-
Statins attenuate antiviral IFN-β and ISG expression via inhibition of IRF3 and JAK/STAT signaling in poly(I:C)-treated hyperlipidemic mice and macrophages.FEBS J. 2021 Jul;288(14):4249-4266. doi: 10.1111/febs.15712. Epub 2021 Feb 4. FEBS J. 2021. PMID: 33452755
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
-
Type I interferon-regulated gene expression and signaling in murine mixed glial cells lacking signal transducers and activators of transcription 1 or 2 or interferon regulatory factor 9.J Biol Chem. 2017 Apr 7;292(14):5845-5859. doi: 10.1074/jbc.M116.756510. Epub 2017 Feb 17. J Biol Chem. 2017. PMID: 28213522 Free PMC article.
-
Signal Integration of IFN-I and IFN-II With TLR4 Involves Sequential Recruitment of STAT1-Complexes and NFκB to Enhance Pro-inflammatory Transcription.Front Immunol. 2019 Jun 4;10:1253. doi: 10.3389/fimmu.2019.01253. eCollection 2019. Front Immunol. 2019. PMID: 31231385 Free PMC article.
-
The JAK-STAT pathway at twenty.Immunity. 2012 Apr 20;36(4):503-14. doi: 10.1016/j.immuni.2012.03.013. Immunity. 2012. PMID: 22520844 Free PMC article. Review.
Cited by
-
Identification of Two Genetic Loci Associated with Leukopenia after Chemotherapy in Patients with Breast Cancer.Clin Cancer Res. 2022 Aug 2;28(15):3342-3355. doi: 10.1158/1078-0432.CCR-20-4774. Clin Cancer Res. 2022. PMID: 35653140 Free PMC article.
-
Mouse Cd59b but not Cd59a is upregulated to protect cells from complement attack in response to inflammatory stimulation.Genes Immun. 2015 Oct;16(7):437-45. doi: 10.1038/gene.2015.29. Epub 2015 Jul 23. Genes Immun. 2015. PMID: 26204229
-
SRF Rearrangements in Soft Tissue Tumors with Muscle Differentiation.Biomolecules. 2022 Nov 12;12(11):1678. doi: 10.3390/biom12111678. Biomolecules. 2022. PMID: 36421692 Free PMC article.
-
Molecular Mechanisms of Leukocyte Migration and Its Potential Targeting-Lessons Learned From MKL1/SRF-Related Primary Immunodeficiency Diseases.Front Immunol. 2021 Feb 22;12:615477. doi: 10.3389/fimmu.2021.615477. eCollection 2021. Front Immunol. 2021. PMID: 33692789 Free PMC article. Review.
-
RNase L attenuates mitogen-stimulated gene expression via transcriptional and post-transcriptional mechanisms to limit the proliferative response.J Biol Chem. 2014 Nov 28;289(48):33629-43. doi: 10.1074/jbc.M114.589556. Epub 2014 Oct 9. J Biol Chem. 2014. PMID: 25301952 Free PMC article.
References
-
- Akira S. Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. - PubMed
-
- Barton GM. Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300(5625):1524–1525. - PubMed
-
- Chai J. Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol. 2002;53(2):147–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

