Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis
- PMID: 23704319
- PMCID: PMC3884067
- DOI: 10.1136/gutjnl-2012-304219
Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis
Abstract
Objective: Hypomethylation of LINE-1 elements has emerged as a distinguishing feature in human cancers. Limited evidence indicates that some LINE-1 elements encode an additional internal antisense promoter, and increased hypomethylation of this region may lead to inadvertent activation of evolutionarily methylation-silenced downstream genes. However, the significance of this fundamental epigenetic mechanism in colorectal cancer (CRC) has not been investigated previously.
Design: We analysed tissue specimens from 77 CRC patients with matched sets of normal colonic mucosa, primary CRC tissues (PC), and liver metastasis tissues (LM). LINE-1 methylation levels were determined by quantitative bisulfite pyrosequencing. MET, RAB3IP and CHRM3 protein expression was determined by western blotting and IHC. MET proto-oncogene transcription and 5-hydroxymethylcytosine (5-hmc) were evaluated by quantitative real-time-PCR.
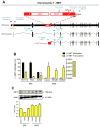
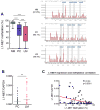
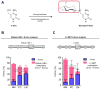
Results: Global LINE-1 methylation levels in LM were significantly lower compared with the matched PC (PC=66.2% vs LM=63.8%; p<0.001). More importantly, we observed that specific LINE-1 sequences residing within the intronic regions of multiple proto-oncogenes, MET (p<0.001), RAB3IP (p=0.05) and CHRM3 (p=0.01), were significantly hypomethylated in LM tissues compared with corresponding matched PC. Furthermore, reduced methylation of specific LINE-1 elements within the MET gene inversely correlated with induction of MET expression in CRC metastases (R=-0.44; p<0.0001). Finally, increased 5-hmc content was associated with LINE-1 hypomethylation.
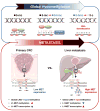
Conclusions: Our results provide novel evidence that hypomethylation of specific LINE-1 elements permits inadvertent activation of methylation-silenced MET, RAB3IP and CHRM3 proto-oncogenes in CRC metastasis. Moreover, since 5-hmc content inversely correlated with LINE-1 hypomethylation in neoplastic tissues, our results provide important mechanistic insights into the fundamental processes underlying global DNA hypomethylation in human CRC.
Keywords: Colorectal Neoplasia; Methylation; Oncogenes.
Conflict of interest statement
Figures


Comment in
-
5-Methylcytosine hydroxylation-mediated LINE-1 hypomethylation: a novel mechanism of proto-oncogenes activation in colorectal cancer?Gut. 2014 Apr;63(4):538-9. doi: 10.1136/gutjnl-2013-305176. Epub 2013 Jun 27. Gut. 2014. PMID: 23812322 Free PMC article. No abstract available.
Similar articles
-
5-Methylcytosine hydroxylation-mediated LINE-1 hypomethylation: a novel mechanism of proto-oncogenes activation in colorectal cancer?Gut. 2014 Apr;63(4):538-9. doi: 10.1136/gutjnl-2013-305176. Epub 2013 Jun 27. Gut. 2014. PMID: 23812322 Free PMC article. No abstract available.
-
LINE-1 is preferentially hypomethylated within adenomatous polyps in the presence of synchronous colorectal cancer.Clin Epigenetics. 2017 Mar 9;9:25. doi: 10.1186/s13148-017-0325-7. eCollection 2017. Clin Epigenetics. 2017. PMID: 28293326 Free PMC article.
-
Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma.Ann Surg Oncol. 2014 Dec;21 Suppl 4:S729-35. doi: 10.1245/s10434-014-3874-4. Epub 2014 Jul 4. Ann Surg Oncol. 2014. PMID: 24992910
-
Long interspersed nucleotide element-1 (LINE-1) methylation in colorectal cancer.Clin Chim Acta. 2019 Jan;488:209-214. doi: 10.1016/j.cca.2018.11.018. Epub 2018 Nov 13. Clin Chim Acta. 2019. PMID: 30445031 Review.
-
LINE-1 methylation level and prognosis in pancreas cancer: pyrosequencing technology and literature review.Surg Today. 2017 Dec;47(12):1450-1459. doi: 10.1007/s00595-017-1539-1. Epub 2017 May 23. Surg Today. 2017. PMID: 28536860 Review.
Cited by
-
Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers.Gastroenterology. 2015 Oct;149(5):1204-1225.e12. doi: 10.1053/j.gastro.2015.07.011. Epub 2015 Jul 26. Gastroenterology. 2015. PMID: 26216839 Free PMC article. Review.
-
SMAD3 Hypomethylation as a Biomarker for Early Prediction of Colorectal Cancer.Int J Mol Sci. 2020 Oct 7;21(19):7395. doi: 10.3390/ijms21197395. Int J Mol Sci. 2020. PMID: 33036415 Free PMC article.
-
Comprehensive identification of onco-exaptation events in bladder cancer cell lines revealed L1PA2-SYT1 as a prognosis-relevant event.iScience. 2023 Nov 17;26(12):108482. doi: 10.1016/j.isci.2023.108482. eCollection 2023 Dec 15. iScience. 2023. PMID: 38058305 Free PMC article.
-
Methylation of LINE-1 in cell-free DNA serves as a liquid biopsy biomarker for human breast cancers and dog mammary tumors.Sci Rep. 2019 Jan 17;9(1):175. doi: 10.1038/s41598-018-36470-5. Sci Rep. 2019. PMID: 30655558 Free PMC article.
-
Activation of LINE-1 Retrotransposon Increases the Risk of Epithelial-Mesenchymal Transition and Metastasis in Epithelial Cancer.Curr Mol Med. 2015;15(7):588-97. doi: 10.2174/1566524015666150831130827. Curr Mol Med. 2015. PMID: 26321759 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
