Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny
- PMID: 23696731
- PMCID: PMC3656100
- DOI: 10.1371/journal.ppat.1003333
Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny
Abstract
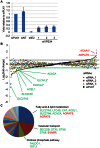
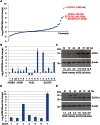
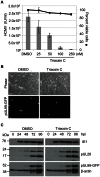
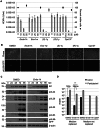
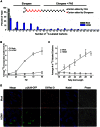
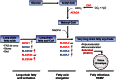
Human cytomegalovirus hijacks host cell metabolism, increasing the flux of carbon from glucose to malonyl-CoA, the committed precursor to fatty acid synthesis and elongation. Inhibition of acetyl-CoA carboxylase blocks the production of progeny virus. To probe further the role of fatty acid metabolism during infection, we performed an siRNA screen to identify host cell metabolic enzymes needed for the production of infectious cytomegalovirus progeny. The screen predicted that multiple long chain acyl-CoA synthetases and fatty acid elongases are needed during infection, and the levels of RNAs encoding several of these enzymes were upregulated by the virus. Roles for acyl-CoA synthetases and elongases during infection were confirmed by using small molecule antagonists. Consistent with a role for these enzymes, mass spectrometry-based fatty acid analysis with ¹³C-labeling revealed that malonyl-CoA is consumed by elongases to produce very long chain fatty acids, generating an approximately 8-fold increase in C26-C34 fatty acid tails in infected cells. The virion envelope was yet further enriched in C26-C34 saturated fatty acids, and elongase inhibitors caused the production of virions with lower levels of these fatty acids and markedly reduced infectivity. These results reveal a dependence of cytomegalovirus on very long chain fatty acid metabolism.
Conflict of interest statement
I have read the journal's policy and have the following conflicts: JDR and TS are shareholders and receive consulting fees from Kadmon Corp. This does not alter our adherence to all PLoS Pathogens policies on sharing data and materials.
Figures

Similar articles
-
Human Cytomegalovirus Uses a Host Stress Response To Balance the Elongation of Saturated/Monounsaturated and Polyunsaturated Very-Long-Chain Fatty Acids.mBio. 2021 May 4;12(3):e00167-21. doi: 10.1128/mBio.00167-21. mBio. 2021. PMID: 33947752 Free PMC article.
-
Human Cytomegalovirus pUL37x1 Is Important for Remodeling of Host Lipid Metabolism.J Virol. 2019 Oct 15;93(21):e00843-19. doi: 10.1128/JVI.00843-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31391267 Free PMC article.
-
Fatty acid elongase 7 catalyzes lipidome remodeling essential for human cytomegalovirus replication.Cell Rep. 2015 Mar 3;10(8):1375-85. doi: 10.1016/j.celrep.2015.02.003. Epub 2015 Feb 26. Cell Rep. 2015. PMID: 25732827 Free PMC article.
-
Role of the malonyl-CoA synthetase ACSF3 in mitochondrial metabolism.Adv Biol Regul. 2019 Jan;71:34-40. doi: 10.1016/j.jbior.2018.09.002. Epub 2018 Sep 5. Adv Biol Regul. 2019. PMID: 30201289 Free PMC article. Review.
-
Malonyl-CoA: the regulator of fatty acid synthesis and oxidation.J Clin Invest. 2012 Jun;122(6):1958-9. doi: 10.1172/jci63967. J Clin Invest. 2012. PMID: 22833869 Free PMC article. Review.
Cited by
-
Metabolic Enzymes in Viral Infection and Host Innate Immunity.Viruses. 2023 Dec 24;16(1):35. doi: 10.3390/v16010035. Viruses. 2023. PMID: 38257735 Free PMC article. Review.
-
Peroxisomal very long-chain fatty acid transport is targeted by herpesviruses and the antiviral host response.Commun Biol. 2022 Sep 9;5(1):944. doi: 10.1038/s42003-022-03867-y. Commun Biol. 2022. PMID: 36085307 Free PMC article.
-
Infection-Induced Peroxisome Biogenesis Is a Metabolic Strategy for Herpesvirus Replication.Cell Host Microbe. 2018 Oct 10;24(4):526-541.e7. doi: 10.1016/j.chom.2018.09.002. Epub 2018 Sep 27. Cell Host Microbe. 2018. PMID: 30269970 Free PMC article.
-
Manipulation of host pathways by human cytomegalovirus: insights from genome-wide studies.Semin Immunopathol. 2014 Nov;36(6):651-8. doi: 10.1007/s00281-014-0443-7. Epub 2014 Sep 27. Semin Immunopathol. 2014. PMID: 25260940 Review.
-
Ketogenic Diet as a Preventive and Supportive Care for COVID-19 Patients.Nutrients. 2021 Mar 20;13(3):1004. doi: 10.3390/nu13031004. Nutrients. 2021. PMID: 33804603 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

