Introns of plant pri-miRNAs enhance miRNA biogenesis
- PMID: 23681439
- PMCID: PMC3701235
- DOI: 10.1038/embor.2013.62
Introns of plant pri-miRNAs enhance miRNA biogenesis
Abstract
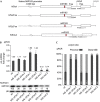
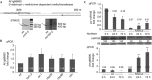
Plant MIR genes are independent transcription units that encode long primary miRNA precursors, which usually contain introns. For two miRNA genes, MIR163 and MIR161, we show that introns are crucial for the accumulation of proper levels of mature miRNA. Removal of the intron in both cases led to a drop-off in the level of mature miRNAs. We demonstrate that the stimulating effects of the intron mostly reside in the 5'ss rather than on a genuine splicing event. Our findings are biologically significant as the presence of functional splice sites in the MIR163 gene appears mandatory for pathogen-triggered accumulation of miR163 and proper regulation of at least one of its targets.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Comment in
-
Feed backwards model for microRNA processing and splicing in plants.EMBO Rep. 2013 Jul;14(7):581-2. doi: 10.1038/embor.2013.77. Epub 2013 Jun 14. EMBO Rep. 2013. PMID: 23764923 Free PMC article. No abstract available.
Similar articles
-
Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs.Nucleic Acids Res. 2009 May;37(9):3083-93. doi: 10.1093/nar/gkp189. Epub 2009 Mar 20. Nucleic Acids Res. 2009. PMID: 19304749 Free PMC article.
-
cis- and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids.Plant Cell. 2011 May;23(5):1729-40. doi: 10.1105/tpc.111.083915. Epub 2011 May 20. Plant Cell. 2011. PMID: 21602291 Free PMC article.
-
Enhanced microRNA accumulation through stemloop-adjacent introns.EMBO Rep. 2013 Jul;14(7):615-21. doi: 10.1038/embor.2013.58. Epub 2013 May 10. EMBO Rep. 2013. PMID: 23661080 Free PMC article.
-
The crosstalk between plant microRNA biogenesis factors and the spliceosome.Plant Signal Behav. 2013 Nov;8(11):e26955. doi: 10.4161/psb.26955. Epub 2013 Dec 3. Plant Signal Behav. 2013. PMID: 24300047 Free PMC article. Review.
-
Posttranscriptional coordination of splicing and miRNA biogenesis in plants.Wiley Interdiscip Rev RNA. 2017 May;8(3). doi: 10.1002/wrna.1403. Epub 2016 Nov 9. Wiley Interdiscip Rev RNA. 2017. PMID: 27863087 Review.
Cited by
-
The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis.Nucleic Acids Res. 2015 Sep 30;43(17):8283-98. doi: 10.1093/nar/gkv751. Epub 2015 Jul 30. Nucleic Acids Res. 2015. PMID: 26227967 Free PMC article.
-
NPK macronutrients and microRNA homeostasis.Front Plant Sci. 2015 Jun 16;6:451. doi: 10.3389/fpls.2015.00451. eCollection 2015. Front Plant Sci. 2015. PMID: 26136763 Free PMC article. Review.
-
Salt Stress Reveals a New Role for ARGONAUTE1 in miRNA Biogenesis at the Transcriptional and Posttranscriptional Levels.Plant Physiol. 2016 Sep;172(1):297-312. doi: 10.1104/pp.16.00830. Epub 2016 Jul 6. Plant Physiol. 2016. PMID: 27385819 Free PMC article.
-
A miRNA-Encoded Small Peptide, vvi-miPEP171d1, Regulates Adventitious Root Formation.Plant Physiol. 2020 Jun;183(2):656-670. doi: 10.1104/pp.20.00197. Epub 2020 Apr 2. Plant Physiol. 2020. PMID: 32241877 Free PMC article.
-
Regulation of pri-MIRNA processing: mechanistic insights into the miRNA homeostasis in plant.Plant Cell Rep. 2021 May;40(5):783-798. doi: 10.1007/s00299-020-02660-7. Epub 2021 Jan 16. Plant Cell Rep. 2021. PMID: 33454802 Review.
References
-
- Voinnet O (2009) Origin, biogenesis and activity of plant microRNAs. Cell 136: 669–687 - PubMed
-
- Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 - PubMed
-
- Kruszka K, Pieczynski M, Windels D, Bielewicz D, Jarmolowski A, Szweykowska-Kulinska Z, Vazquez F (2012) Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol 169: 1664–1672 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

