Quantitative proteomics reveals the temperature-dependent proteins encoded by a series of cluster genes in thermoanaerobacter tengcongensis
- PMID: 23665590
- PMCID: PMC3734584
- DOI: 10.1074/mcp.M112.025817
Quantitative proteomics reveals the temperature-dependent proteins encoded by a series of cluster genes in thermoanaerobacter tengcongensis
Abstract
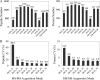
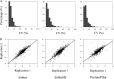
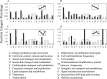
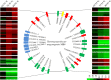
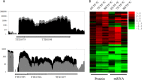
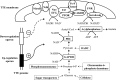
Comprehensive and quantitative information of the thermophile proteome is an important source for understanding of the survival mechanism under high growth temperature. Thermoanaerobacter tengcongensis (T. tengcongensis), a typical anaerobic thermophilic eubacterium, was selected to quantitatively evaluate its protein abundance changes in response to four different temperatures. With optimized procedures of isobaric tags for relative and absolute quantitation quantitative proteomics (iTRAQ), such as peptide fractionation with high-pH reverse phase (RP) high performance liquid chromatography (HPLC), tandem MS acquisition mode in LTQ Orbitrap Velos MS, and evaluation of the quantification algorithms, high quality of the quantitative information of the peptides identified were acquired. In total, 1589 unique proteins were identified and defined 251 as the temperature-dependent proteins. Analysis of genomic locations toward the correspondent genes of these temperature-dependent proteins revealed that more than 30% were contiguous units with relevant biological functions, which are likely to form the operon structures in T. tengcongensis. The RNA sequencing (RNA-seq) data further demonstrated that these cluster genes were cotranscribed, and their mRNA abundance changes responding to temperature exhibited the similar trends as the proteomic results, suggesting that the temperature-dependent proteins are highly associated with the correspondent transcription status. Hence, the operon regulation is likely an energy-efficient mode for T. tengcongensis survival. In addition, evaluation to the functions of differential proteomes indicated that the abundance of the proteins participating in sulfur-respiration on the plasma membrane was decreased as the temperature increased, whereas the glycolysis-related protein abundance was increased. The energy supply in T. tengcongensis at high temperature is, therefore, speculated not mainly through the respiration chain reactions.
Figures
Similar articles
-
[Dynamic analysis of the heat shock protein 60 and its gene expression in Thermoanaerobacter tengcongensis induced by temperature increase].Wei Sheng Wu Xue Bao. 2006 Apr;46(2):269-74. Wei Sheng Wu Xue Bao. 2006. PMID: 16736590 Chinese.
-
Proteomic analysis on the temperature-dependent complexes in Thermoanaerobacter tengcongensis.Proteomics. 2009 Jun;9(11):3189-200. doi: 10.1002/pmic.200800650. Proteomics. 2009. PMID: 19526551
-
The proteomic alterations of Thermoanaerobacter tengcongensis cultured at different temperatures.Proteomics. 2007 May;7(9):1409-19. doi: 10.1002/pmic.200500226. Proteomics. 2007. PMID: 17469076
-
Proteomics technologies for the global identification and quantification of proteins.Adv Protein Chem Struct Biol. 2010;80:1-44. doi: 10.1016/B978-0-12-381264-3.00001-1. Adv Protein Chem Struct Biol. 2010. PMID: 21109216 Review.
-
Global relative and absolute quantitation in microbial proteomics.Curr Opin Microbiol. 2012 Jun;15(3):364-72. doi: 10.1016/j.mib.2012.02.005. Epub 2012 Mar 21. Curr Opin Microbiol. 2012. PMID: 22445110 Review.
Cited by
-
Visualization of proteomics data using R and bioconductor.Proteomics. 2015 Apr;15(8):1375-89. doi: 10.1002/pmic.201400392. Proteomics. 2015. PMID: 25690415 Free PMC article. Review.
-
High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.Nat Commun. 2022 Jan 25;13(1):476. doi: 10.1038/s41467-022-27967-9. Nat Commun. 2022. PMID: 35079002 Free PMC article.
-
Proteomics analyses for the global proteins in the brain tissues of different human prion diseases.Mol Cell Proteomics. 2015 Apr;14(4):854-69. doi: 10.1074/mcp.M114.038018. Epub 2015 Jan 23. Mol Cell Proteomics. 2015. PMID: 25616867 Free PMC article.
-
High level of serum AMBP is associated with poor response to paclitaxel-capecitabine chemotherapy in advanced gastric cancer patients.Med Oncol. 2013 Dec;30(4):748. doi: 10.1007/s12032-013-0748-8. Epub 2013 Oct 18. Med Oncol. 2013. PMID: 24135868
-
Thermal stability of glucokinases in Thermoanaerobacter tengcongensis.Biomed Res Int. 2013;2013:646539. doi: 10.1155/2013/646539. Epub 2013 Aug 24. Biomed Res Int. 2013. PMID: 24058911 Free PMC article.
References
-
- Cannio R., Rossi M., Bartolucci S. (1994) A few amino acid substitutions are responsible for the higher thermostability of a novel NAD(+)-dependent bacillar alcohol dehydrogenase. Eur. J. Biochem. 222, 345–352 - PubMed
-
- Arnórsdóttir J., Sigtryggsdottir A. R., Thorbjarnardóttir S. H., Kristjánsson M. M. (2009) Effect of proline substitutions on stability and kinetic properties of a cold adapted subtilase. J. Biochem. 145, 325–329 - PubMed
-
- Chen J., Stites W. E. (2004) Replacement of staphylococcal nuclease hydrophobic core residues with those from thermophilic homologues indicates packing is improved in some thermostable proteins. J. Mol. Biol. 344, 271–280 - PubMed
-
- Bezsudnova E. Y., Boyko K. M., Polyakov K. M., Dorovatovskiy P. V., Stekhanova T. N., Gumerov V. M., Ravin N. V., Skryabin K. G., Kovalchuk M. V., Popov V. O. (2012) Structural insight into the molecular basis of polyextremophilicity of short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Thermococcus sibiricus. Biochimie, 94, 2628–2638 - PubMed
-
- Hakulinen N., Turunen O., Jänis J., Leisola M., Rouvinen J. (2003) Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability. Eur. J. Biochem. 270, 1399–1412 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

