Structure of the human smoothened receptor bound to an antitumour agent
- PMID: 23636324
- PMCID: PMC3657389
- DOI: 10.1038/nature12167
Structure of the human smoothened receptor bound to an antitumour agent
Abstract
The smoothened (SMO) receptor, a key signal transducer in the hedgehog signalling pathway, is responsible for the maintenance of normal embryonic development and is implicated in carcinogenesis. It is classified as a class frizzled (class F) G-protein-coupled receptor (GPCR), although the canonical hedgehog signalling pathway involves the GLI transcription factors and the sequence similarity with class A GPCRs is less than 10%. Here we report the crystal structure of the transmembrane domain of the human SMO receptor bound to the small-molecule antagonist LY2940680 at 2.5 Å resolution. Although the SMO receptor shares the seven-transmembrane helical fold, most of the conserved motifs for class A GPCRs are absent, and the structure reveals an unusually complex arrangement of long extracellular loops stabilized by four disulphide bonds. The ligand binds at the extracellular end of the seven-transmembrane-helix bundle and forms extensive contacts with the loops.
Figures
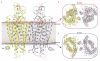
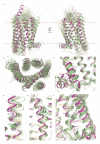
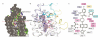
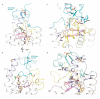

Comment in
-
G protein-coupled receptors: pioneering Frizzled family receptor structure solved.Nat Rev Drug Discov. 2013 Jun;12(6):424. doi: 10.1038/nrd4030. Epub 2013 May 17. Nat Rev Drug Discov. 2013. PMID: 23681005 No abstract available.
Similar articles
-
Molecular modeling study on the dynamical structural features of human smoothened receptor and binding mechanism of antagonist LY2940680 by metadynamics simulation and free energy calculation.Biochim Biophys Acta. 2014 Jul;1840(7):2128-38. doi: 10.1016/j.bbagen.2014.03.010. Epub 2014 Mar 15. Biochim Biophys Acta. 2014. PMID: 24637074
-
[Structure of the Smoothened receptor].Med Sci (Paris). 2013 Oct;29(10):855-60. doi: 10.1051/medsci/20132910012. Epub 2013 Oct 18. Med Sci (Paris). 2013. PMID: 24148123 Review. French.
-
Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling.Nat Commun. 2013;4:2965. doi: 10.1038/ncomms3965. Nat Commun. 2013. PMID: 24351982 Free PMC article.
-
Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.Nat Commun. 2014 Jul 10;5:4355. doi: 10.1038/ncomms5355. Nat Commun. 2014. PMID: 25008467 Free PMC article.
-
Structural and Druggability Landscape of Frizzled G Protein-Coupled Receptors.Trends Biochem Sci. 2018 Dec;43(12):1033-1046. doi: 10.1016/j.tibs.2018.09.002. Epub 2018 Oct 8. Trends Biochem Sci. 2018. PMID: 30309741 Review.
Cited by
-
Residues remote from the binding pocket control the antagonist selectivity towards the corticotropin-releasing factor receptor-1.Sci Rep. 2015 Jan 28;5:8066. doi: 10.1038/srep08066. Sci Rep. 2015. PMID: 25628267 Free PMC article.
-
Generic GPCR residue numbers - aligning topology maps while minding the gaps.Trends Pharmacol Sci. 2015 Jan;36(1):22-31. doi: 10.1016/j.tips.2014.11.001. Epub 2014 Dec 22. Trends Pharmacol Sci. 2015. PMID: 25541108 Free PMC article. Review.
-
A Recurrent Mosaic Mutation in SMO, Encoding the Hedgehog Signal Transducer Smoothened, Is the Major Cause of Curry-Jones Syndrome.Am J Hum Genet. 2016 Jun 2;98(6):1256-1265. doi: 10.1016/j.ajhg.2016.04.007. Epub 2016 May 26. Am J Hum Genet. 2016. PMID: 27236920 Free PMC article.
-
Structure of class B GPCR corticotropin-releasing factor receptor 1.Nature. 2013 Jul 25;499(7459):438-43. doi: 10.1038/nature12357. Epub 2013 Jul 17. Nature. 2013. PMID: 23863939
-
Identification of Novel Smoothened Ligands Using Structure-Based Docking.PLoS One. 2016 Aug 4;11(8):e0160365. doi: 10.1371/journal.pone.0160365. eCollection 2016. PLoS One. 2016. PMID: 27490099 Free PMC article.
References
-
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi:10.1101/gad.938601. - PubMed
-
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi:10.1038/384176a0. - PubMed
-
- Stone DM, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi:10.1038/384129a0. - PubMed
-
- Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi:10.1038/nature00989. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

