Unusual fragmentation pathways in collagen glycopeptides
- PMID: 23633013
- PMCID: PMC3679267
- DOI: 10.1007/s13361-013-0624-y
Unusual fragmentation pathways in collagen glycopeptides
Abstract
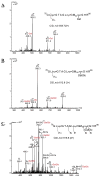
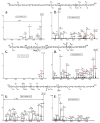
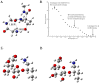
Collagens are the most abundant glycoproteins in the body. One characteristic of this protein family is that the amino acid sequence consists of repeats of three amino acids -(X-Y-Gly)n. Within this motif, the Y residue is often 4-hydroxyproline (HyP) or 5-hydroxylysine (HyK). Glycosylation in collagen occurs at the 5-OH group in HyK in the form of two glycosides, galactosylhydroxylysine (Gal-HyK) and glucosyl galactosylhydroxylysine (GlcGal-HyK). In collision induced dissociation (CID), collagen tryptic glycopeptides exhibit unexpected gas-phase dissociation behavior compared to typical N- and O-linked glycopeptides (i.e., in addition to glycosidic bond cleavages, extensive cleavages of the amide bonds are observed). The Gal- or GlcGal- glycan modifications are largely retained on the fragment ions. These features enable unambiguous determination of the amino acid sequence of collagen glycopeptides and the location of the glycosylation site. This dissociation pattern was consistent for all analyzed collagen glycopeptides, regardless of their length or amino acid composition, collagen type or tissue. The two fragmentation pathways-amide bond and glycosidic bond cleavage-are highly competitive in collagen tryptic glycopeptides. The number of ionizing protons relative to the number of basic sites (i.e., Arg, Lys, HyK, and N-terminus) is a major driving force of the fragmentation. We present here our experimental results and employ quantum mechanics calculations to understand the factors enhancing the labile character of the amide bonds and the stability of hydroxylysine glycosides in gas phase dissociation of collagen glycopeptides.
Figures

Similar articles
-
Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry.Mol Cell Proteomics. 2012 Jun;11(6):M111.010397. doi: 10.1074/mcp.M111.010397. Epub 2012 Jan 13. Mol Cell Proteomics. 2012. PMID: 22247541 Free PMC article.
-
Parallel Determination of Polypeptide and Oligosaccharide Connectivities by Energy-Resolved Collison-Induced Dissociation of Protonated O-Glycopeptides Derived from Nonspecific Proteolysis.J Am Soc Mass Spectrom. 2020 Mar 4;31(3):624-632. doi: 10.1021/jasms.9b00065. Epub 2020 Feb 21. J Am Soc Mass Spectrom. 2020. PMID: 32126781 Free PMC article.
-
Sialic acid capture-and-release and LC-MS(n) analysis of glycopeptides.Methods Mol Biol. 2013;951:79-100. doi: 10.1007/978-1-62703-146-2_7. Methods Mol Biol. 2013. PMID: 23296526
-
Glycoproteomics based on tandem mass spectrometry of glycopeptides.J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1-2):115-28. doi: 10.1016/j.jchromb.2006.09.041. Epub 2006 Oct 17. J Chromatogr B Analyt Technol Biomed Life Sci. 2007. PMID: 17049937 Review.
-
Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides.Glycoconj J. 2016 Jun;33(3):261-72. doi: 10.1007/s10719-016-9649-3. Epub 2016 Jan 18. Glycoconj J. 2016. PMID: 26780731 Review.
Cited by
-
Zebrafish Tric-b is required for skeletal development and bone cells differentiation.Front Endocrinol (Lausanne). 2023 Jan 23;14:1002914. doi: 10.3389/fendo.2023.1002914. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36755921 Free PMC article.
-
Assessment of Collagen in Translational Models of Lung Research.Adv Exp Med Biol. 2023;1413:213-244. doi: 10.1007/978-3-031-26625-6_11. Adv Exp Med Biol. 2023. PMID: 37195533
-
Comprehensive Characterization of Glycosylation and Hydroxylation of Basement Membrane Collagen IV by High-Resolution Mass Spectrometry.J Proteome Res. 2016 Jan 4;15(1):245-58. doi: 10.1021/acs.jproteome.5b00767. Epub 2015 Dec 9. J Proteome Res. 2016. PMID: 26593852 Free PMC article.
-
Discovery and characterization of hydroxylysine in recombinant monoclonal antibodies.MAbs. 2016;8(2):371-8. doi: 10.1080/19420862.2015.1122148. Epub 2015 Dec 14. MAbs. 2016. PMID: 26651858 Free PMC article.
-
Glycosylation and cross-linking in bone type I collagen.J Biol Chem. 2014 Aug 15;289(33):22636-22647. doi: 10.1074/jbc.M113.528513. Epub 2014 Jun 23. J Biol Chem. 2014. PMID: 24958722 Free PMC article.
References
-
- Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. 2009 - PubMed
-
- Settineri CA, Medzihradszky KF, Masiarz FR, Burlingame AL, Chu C, George-Nascimento C. Characterization of O-glycosylation sites in recombinant B-chain of platelet-derived growth factor expressed in yeast using liquid secondary ion mass spectrometry, tandem mass spectrometry and Edman sequence analysis. Biomed Environ Mass Spectrom. 1990;19(11):665–76. - PubMed
-
- Medzihradszky KF, Gillece-Castro BL, Hardy MR, Townsend RR, Burlingame AL. Structural Elucidation of O-linked Glycopeptides by High Energy Collision-Induced Dissociation. Journal of the American Society for Mass Spectrometry. 1996;7:319–328. - PubMed
-
- Hanisch FG, Green BN, Bateman R, Peter-Katalinic J. Localization of O-glycosylation sites of MUC1 tandem repeats by QTOF ESI mass spectrometry. J Mass Spectrom. 1998;33(4):358–62. - PubMed
-
- Alving K, Paulsen H, Peter-Katalinic J. Characterization of O-glycosylation sites in MUC2 glycopeptides by nanoelectrospray QTOF mass spectrometry. J Mass Spectrom. 1999;34(4):395–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

