Predicting hotspots for influenza virus reassortment
- PMID: 23628436
- PMCID: PMC3647410
- DOI: 10.3201/eid1904.120903
Predicting hotspots for influenza virus reassortment
Abstract
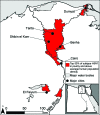
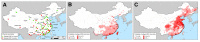
The 1957 and 1968 influenza pandemics, each of which killed ≈1 million persons, arose through reassortment events. Influenza virus in humans and domestic animals could reassort and cause another pandemic. To identify geographic areas where agricultural production systems are conducive to reassortment, we fitted multivariate regression models to surveillance data on influenza A virus subtype H5N1 among poultry in China and Egypt and subtype H3N2 among humans. We then applied the models across Asia and Egypt to predict where subtype H3N2 from humans and subtype H5N1 from birds overlap; this overlap serves as a proxy for co-infection and in vivo reassortment. For Asia, we refined the prioritization by identifying areas that also have high swine density. Potential geographic foci of reassortment include the northern plains of India, coastal and central provinces of China, the western Korean Peninsula and southwestern Japan in Asia, and the Nile Delta in Egypt.
Keywords: avian influenza; influenza; influenza A virus H3N2 subtype; influenza A virus H5N1 subtype; influenza in birds; reassortant viruses; viruses; zoonoses.
Figures

Similar articles
-
Reassortant between human-Like H3N2 and avian H5 subtype influenza A viruses in pigs: a potential public health risk.PLoS One. 2010 Sep 7;5(9):e12591. doi: 10.1371/journal.pone.0012591. PLoS One. 2010. PMID: 20830295 Free PMC article.
-
Evidence of infection with avian, human, and swine influenza viruses in pigs in Cairo, Egypt.Arch Virol. 2018 Feb;163(2):359-364. doi: 10.1007/s00705-017-3619-3. Epub 2017 Oct 26. Arch Virol. 2018. PMID: 29075888
-
Reassortment between Avian H5N1 and human influenza viruses is mainly restricted to the matrix and neuraminidase gene segments.PLoS One. 2013;8(3):e59889. doi: 10.1371/journal.pone.0059889. Epub 2013 Mar 20. PLoS One. 2013. PMID: 23527283 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Epidemiology, ecology and gene pool of influenza A virus in Egypt: will Egypt be the epicentre of the next influenza pandemic?Virulence. 2015;6(1):6-18. doi: 10.4161/21505594.2014.992662. Virulence. 2015. PMID: 25635701 Free PMC article. Review.
Cited by
-
Complete Genome Sequence of a Novel Reassortant Avian Influenza H1N2 Virus Isolated from a Domestic Sparrow in 2012.Genome Announc. 2013 Jul 18;1(4):e00431-13. doi: 10.1128/genomeA.00431-13. Genome Announc. 2013. PMID: 23868121 Free PMC article.
-
Nine challenges in modelling the emergence of novel pathogens.Epidemics. 2015 Mar;10:35-9. doi: 10.1016/j.epidem.2014.09.002. Epub 2014 Sep 19. Epidemics. 2015. PMID: 25843380 Free PMC article.
-
Avian Influenza A Virus Infection among Workers at Live Poultry Markets, China, 2013-2016.Emerg Infect Dis. 2018 Jul;24(7):1246-1256. doi: 10.3201/eid2407.172059. Emerg Infect Dis. 2018. PMID: 29912708 Free PMC article.
-
Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia.Nat Commun. 2014 Jun 17;5:4116. doi: 10.1038/ncomms5116. Nat Commun. 2014. PMID: 24937647 Free PMC article.
-
A global-scale ecological niche model to predict SARS-CoV-2 coronavirus infection rate.Ecol Modell. 2020 Sep 1;431:109187. doi: 10.1016/j.ecolmodel.2020.109187. Epub 2020 Jun 20. Ecol Modell. 2020. PMID: 32834369 Free PMC article.
References
-
- Turner PE. Parasitism between co-infecting bacteriophages. Adv Ecol Res. 2005;37:309–32. 10.1016/S0065-2504(04)37010-8 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
