Informatics of protein and posttranslational modification detection via shotgun proteomics
- PMID: 23625403
- PMCID: PMC4012295
- DOI: 10.1007/978-1-62703-360-2_14
Informatics of protein and posttranslational modification detection via shotgun proteomics
Abstract
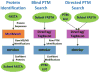
Frequently, proteomic LC-MS/MS data may contain sets of modifications that evade identification during standard database search. For many laboratories, the standard technique to seek posttranslational modifications (PTMs) adds a short list of specified mass shifts to database search configuration. This technique provides information for only the specified PTMs, takes substantial time to run, and drives false discoveries upward through an exponential expansion of search space. This protocol describes a more structured approach to blind PTM discovery through reducing protein lists, targeting attention to a data-driven list of mass shifts, and seeking the resulting short list of modifications through targeted search.
Figures
Similar articles
-
PTMSearchPlus: software tool for automated protein identification and post-translational modification characterization by integrating accurate intact protein mass and bottom-up mass spectrometric data searches.Anal Chem. 2009 Oct 15;81(20):8387-95. doi: 10.1021/ac901163c. Anal Chem. 2009. PMID: 19775152
-
Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics.Adv Exp Med Biol. 2016;919:345-382. doi: 10.1007/978-3-319-41448-5_17. Adv Exp Med Biol. 2016. PMID: 27975226 Review.
-
Software eyes for protein post-translational modifications.Mass Spectrom Rev. 2015 Mar-Apr;34(2):133-47. doi: 10.1002/mas.21425. Epub 2014 Jun 2. Mass Spectrom Rev. 2015. PMID: 24889695 Review.
-
Hunting for unexpected post-translational modifications by spectral library searching with tier-wise scoring.J Proteome Res. 2014 May 2;13(5):2262-71. doi: 10.1021/pr401006g. Epub 2014 Apr 2. J Proteome Res. 2014. PMID: 24661115
-
Human Proteomic Variation Revealed by Combining RNA-Seq Proteogenomics and Global Post-Translational Modification (G-PTM) Search Strategy.J Proteome Res. 2016 Mar 4;15(3):800-8. doi: 10.1021/acs.jproteome.5b00817. Epub 2016 Jan 12. J Proteome Res. 2016. PMID: 26704769 Free PMC article.
Cited by
-
Proteomics and Its Current Application in Biomedical Area: Concise Review.ScientificWorldJournal. 2024 Feb 17;2024:4454744. doi: 10.1155/2024/4454744. eCollection 2024. ScientificWorldJournal. 2024. PMID: 38404932 Free PMC article. Review.
-
Implementation of multiomic mass spectrometry approaches for the evaluation of human health following environmental exposure.Mol Omics. 2024 Jun 10;20(5):296-321. doi: 10.1039/d3mo00214d. Mol Omics. 2024. PMID: 38623720 Free PMC article. Review.
-
Precision diagnostics in children.Camb Prism Precis Med. 2023 Feb 3;1:e17. doi: 10.1017/pcm.2023.4. eCollection 2023. Camb Prism Precis Med. 2023. PMID: 38550930 Free PMC article. Review.
-
Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells.Anal Chem. 2015 Mar 3;87(5):2535-41. doi: 10.1021/ac504685y. Epub 2015 Feb 9. Anal Chem. 2015. PMID: 25654326 Free PMC article.
-
Comprehensive Characterization of Glycosylation and Hydroxylation of Basement Membrane Collagen IV by High-Resolution Mass Spectrometry.J Proteome Res. 2016 Jan 4;15(1):245-58. doi: 10.1021/acs.jproteome.5b00767. Epub 2015 Dec 9. J Proteome Res. 2016. PMID: 26593852 Free PMC article.
References
-
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. - PubMed
-
- Yates JR, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. - PubMed
-
- Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17:2310–2316. - PubMed
-
- Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous