In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation
- PMID: 23610125
- PMCID: PMC3646490
- DOI: 10.1084/jem.20120950
In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation
Erratum in
-
Correction: In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation.J Exp Med. 2017 May 1;214(5):1557. doi: 10.1084/jem.2012095001172017c. Epub 2017 Jan 19. J Exp Med. 2017. PMID: 28104811 Free PMC article. No abstract available.
Expression of concern in
-
Expression of Concern: In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation.J Exp Med. 2022 Nov 7;219(11):e2012095009272022e. doi: 10.1084/jem.2012095009272022e. Epub 2022 Oct 3. J Exp Med. 2022. PMID: 36191251 Free PMC article. No abstract available.
Abstract
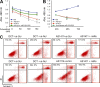
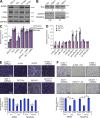
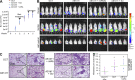
Numerous studies have described the altered expression and the causal role of microRNAs (miRNAs) in human cancer. However, to date, efforts to modulate miRNA levels for therapeutic purposes have been challenging to implement. Here we find that nucleolin (NCL), a major nucleolar protein, posttranscriptionally regulates the expression of a specific subset of miRNAs, including miR-21, miR-221, miR-222, and miR-103, that are causally involved in breast cancer initiation, progression, and drug resistance. We also show that NCL is commonly overexpressed in human breast tumors and that its expression correlates with that of NCL-dependent miRNAs. Finally, inhibition of NCL using guanosine-rich aptamers reduces the levels of NCL-dependent miRNAs and their target genes, thus reducing breast cancer cell aggressiveness both in vitro and in vivo. These findings illuminate a path to novel therapeutic approaches based on NCL-targeting aptamers for the modulation of miRNA expression in the treatment of breast cancer.
Figures


Similar articles
-
Regulation of breast cancer and bone metastasis by microRNAs.Dis Markers. 2013;35(5):369-87. doi: 10.1155/2013/451248. Epub 2013 Sep 26. Dis Markers. 2013. PMID: 24191129 Free PMC article. Review.
-
CRYβB2 enhances tumorigenesis through upregulation of nucleolin in triple negative breast cancer.Oncogene. 2021 Sep;40(38):5752-5763. doi: 10.1038/s41388-021-01975-3. Epub 2021 Aug 2. Oncogene. 2021. PMID: 34341513 Free PMC article.
-
MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway.Tumour Biol. 2016 Jul;37(7):8993-9000. doi: 10.1007/s13277-015-4513-9. Epub 2016 Jan 12. Tumour Biol. 2016. PMID: 26758430
-
Downregulation of miRNA-141 in breast cancer cells is associated with cell migration and invasion: involvement of ANP32E targeting.Cancer Med. 2017 Mar;6(3):662-672. doi: 10.1002/cam4.1024. Epub 2017 Feb 21. Cancer Med. 2017. PMID: 28220627 Free PMC article.
-
Nucleolin‑based targeting strategies in cancer treatment: Focus on cancer immunotherapy (Review).Int J Mol Med. 2023 Sep;52(3):81. doi: 10.3892/ijmm.2023.5284. Epub 2023 Jul 21. Int J Mol Med. 2023. PMID: 37477132 Free PMC article. Review.
Cited by
-
Cellular localization of nucleolin determines the prognosis in cancers: a meta-analysis.J Mol Med (Berl). 2022 Aug;100(8):1145-1157. doi: 10.1007/s00109-022-02228-w. Epub 2022 Jul 21. J Mol Med (Berl). 2022. PMID: 35861882 Free PMC article. Review.
-
SUN2: A potential therapeutic target in cancer.Oncol Lett. 2019 Feb;17(2):1401-1408. doi: 10.3892/ol.2018.9764. Epub 2018 Nov 27. Oncol Lett. 2019. PMID: 30675193 Free PMC article. Review.
-
Regulation of breast cancer and bone metastasis by microRNAs.Dis Markers. 2013;35(5):369-87. doi: 10.1155/2013/451248. Epub 2013 Sep 26. Dis Markers. 2013. PMID: 24191129 Free PMC article. Review.
-
Post-transcriptional regulation of MMP2 mRNA by its interaction with miR-20a and Nucleolin in breast cancer cell lines.Mol Biol Rep. 2021 Mar;48(3):2315-2324. doi: 10.1007/s11033-021-06261-9. Epub 2021 Mar 31. Mol Biol Rep. 2021. PMID: 33788053
-
miRNAs regulated by estrogens, tamoxifen, and endocrine disruptors and their downstream gene targets.Mol Cell Endocrinol. 2015 Dec 15;418 Pt 3(0 3):273-97. doi: 10.1016/j.mce.2015.01.035. Epub 2015 Feb 3. Mol Cell Endocrinol. 2015. PMID: 25659536 Free PMC article. Review.
References
-
- Abdelmohsen K., Tominaga K., Lee E.K., Srikantan S., Kang M.J., Kim M.M., Selimyan R., Martindale J.L., Yang X., Carrier F., et al. 2011. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 39:8513–8530. 10.1093/nar/gkr488 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

