Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway
- PMID: 23603116
- PMCID: PMC3665649
- DOI: 10.1016/j.molcel.2013.03.018
Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway
Abstract
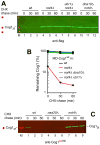
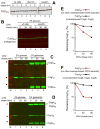
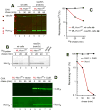
N(α)-terminal acetylation of cellular proteins was recently discovered to create specific degradation signals termed Ac/N-degrons and targeted by the Ac/N-end rule pathway. We show that Hcn1, a subunit of the APC/C ubiquitin ligase, contains an Ac/N-degron that is repressed by Cut9, another APC/C subunit and the ligand of Hcn1. Cog1, a subunit of the Golgi-associated COG complex, is also shown to contain an Ac/N-degron. Cog2 and Cog3, direct ligands of Cog1, can repress this degron. The subunit decoy technique was used to show that the long-lived endogenous Cog1 is destabilized and destroyed via its activated (unshielded) Ac/N-degron if the total level of Cog1 increased in a cell. Hcn1 and Cog1 are the first examples of protein regulation through the physiologically relevant transitions that shield and unshield natural Ac/N-degrons. This mechanistically straightforward circuit can employ the demonstrated conditionality of Ac/N-degrons to regulate subunit stoichiometries and other aspects of protein quality control.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates.Mol Biol Cell. 2013 Apr;24(7):890-900. doi: 10.1091/mbc.E12-11-0838. Epub 2013 Jan 30. Mol Biol Cell. 2013. PMID: 23363603 Free PMC article.
-
Molecular organization of the COG vesicle tethering complex.Nat Struct Mol Biol. 2010 Nov;17(11):1292-7. doi: 10.1038/nsmb.1917. Epub 2010 Oct 24. Nat Struct Mol Biol. 2010. PMID: 20972446 Free PMC article.
-
Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway.Science. 2015 Mar 13;347(6227):1249-1252. doi: 10.1126/science.aaa3844. Science. 2015. PMID: 25766235 Free PMC article.
-
N-Terminal Acetylation-Targeted N-End Rule Proteolytic System: The Ac/N-End Rule Pathway.Mol Cells. 2016 Mar;39(3):169-78. doi: 10.14348/molcells.2016.2329. Epub 2016 Feb 16. Mol Cells. 2016. PMID: 26883906 Free PMC article. Review.
-
The N-end rule pathway and regulation by proteolysis.Protein Sci. 2011 Aug;20(8):1298-345. doi: 10.1002/pro.666. Protein Sci. 2011. PMID: 21633985 Free PMC article. Review.
Cited by
-
Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots.Proteomics. 2015 Jul;15(14):2447-57. doi: 10.1002/pmic.201400530. Epub 2015 Apr 17. Proteomics. 2015. PMID: 25728785 Free PMC article.
-
N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects.Proteomics. 2015 Jul;15(14):2385-401. doi: 10.1002/pmic.201400619. Epub 2015 Jun 16. Proteomics. 2015. PMID: 25914051 Free PMC article. Review.
-
Analyzing N-terminal Arginylation through the Use of Peptide Arrays and Degradation Assays.J Biol Chem. 2016 Sep 30;291(40):20976-20992. doi: 10.1074/jbc.M116.747956. Epub 2016 Aug 10. J Biol Chem. 2016. PMID: 27510035 Free PMC article.
-
Timing and specificity of cotranslational nascent protein modification in bacteria.Proc Natl Acad Sci U S A. 2019 Nov 12;116(46):23050-23060. doi: 10.1073/pnas.1912264116. Epub 2019 Oct 30. Proc Natl Acad Sci U S A. 2019. PMID: 31666319 Free PMC article.
-
Control of protein degradation by N-terminal acetylation and the N-end rule pathway.Exp Mol Med. 2018 Jul 27;50(7):1-8. doi: 10.1038/s12276-018-0097-y. Exp Mol Med. 2018. PMID: 30054456 Free PMC article. Review.
References
-
- Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast to humans. Proc Natl Acad Sci USA. 2009;106:8157–8162. - PMC - PubMed
-
- Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C) Q Rev Biophys. 2011;44:153–190. - PubMed
-
- Collart M, Panasenko O, Nikolaev S. The Not3/5 subunit of the Ccr4-Not complex: A central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal. 2012;25:743–751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

