Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming
- PMID: 23602153
- PMCID: PMC3657391
- DOI: 10.1016/j.cell.2013.04.001
Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming
Abstract
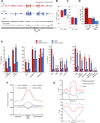
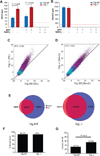
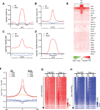
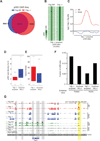
TET proteins oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). 5fC and 5caC are excised by mammalian DNA glycosylase TDG, implicating 5mC oxidation in DNA demethylation. Here, we show that the genomic locations of 5fC can be determined by coupling chemical reduction with biotin tagging. Genome-wide mapping of 5fC in mouse embryonic stem cells (mESCs) reveals that 5fC preferentially occurs at poised enhancers among other gene regulatory elements. Application to Tdg null mESCs further suggests that 5fC production coordinates with p300 in remodeling epigenetic states of enhancers. This process, which is not influenced by 5hmC, appears to be associated with further oxidation of 5hmC and commitment to demethylation through 5fC. Finally, we resolved 5fC at base resolution by hydroxylamine-based protection from bisulfite-mediated deamination, thereby confirming sites of 5fC accumulation. Our results reveal roles of active 5mC/5hmC oxidation and TDG-mediated demethylation in epigenetic tuning at regulatory elements.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Deep C diving: mapping the low-abundance modifications of the DNA demethylation pathway.Genome Biol. 2013 May 29;14(5):118. doi: 10.1186/gb-2013-14-5-118. Genome Biol. 2013. PMID: 23732041 Free PMC article.
-
Dynamics of DNA demethylation.Nat Methods. 2013 Jun;10(6):466. doi: 10.1038/nmeth.2506. Nat Methods. 2013. PMID: 23866334 No abstract available.
Similar articles
-
Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics.Cell. 2013 Apr 25;153(3):692-706. doi: 10.1016/j.cell.2013.04.002. Epub 2013 Apr 18. Cell. 2013. PMID: 23602152 Free PMC article.
-
Whole-Genome Mapping of Epigenetic Modification of 5-Formylcytosine at Single-Base Resolution by Chemical Labeling Enrichment and Deamination Sequencing.Anal Chem. 2024 Mar 19;96(11):4726-4735. doi: 10.1021/acs.analchem.4c00425. Epub 2024 Mar 7. Anal Chem. 2024. PMID: 38450632
-
Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase.Genome Biol. 2012 Aug 17;13(8):R69. doi: 10.1186/gb-2012-13-8-r69. Genome Biol. 2012. PMID: 22902005 Free PMC article.
-
Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review.
-
[Oxidation and deamination of nucleobases as an epigenetic tool].Postepy Hig Med Dosw (Online). 2012 May 24;66:275-86. doi: 10.5604/17322693.997954. Postepy Hig Med Dosw (Online). 2012. PMID: 22706113 Review. Polish.
Cited by
-
Epigenetic Changes in Diabetes and Cardiovascular Risk.Circ Res. 2016 May 27;118(11):1706-22. doi: 10.1161/CIRCRESAHA.116.306819. Circ Res. 2016. PMID: 27230637 Free PMC article. Review.
-
Are there specific readers of oxidized 5-methylcytosine bases?Bioessays. 2016 Oct;38(10):1038-47. doi: 10.1002/bies.201600126. Epub 2016 Aug 2. Bioessays. 2016. PMID: 27480808 Free PMC article. Review.
-
Hydroxymethylation is uniquely distributed within term placenta, and is associated with gene expression.FASEB J. 2016 Aug;30(8):2874-84. doi: 10.1096/fj.201600310R. Epub 2016 Apr 26. FASEB J. 2016. PMID: 27118675 Free PMC article.
-
Active DNA demethylation-The epigenetic gatekeeper of development, immunity, and cancer.Adv Genet (Hoboken). 2020 Nov 27;2(1):e10033. doi: 10.1002/ggn2.10033. eCollection 2021 Mar. Adv Genet (Hoboken). 2020. PMID: 36618446 Free PMC article. Review.
-
5-Formylcytosine-induced DNA-peptide cross-links reduce transcription efficiency, but do not cause transcription errors in human cells.J Biol Chem. 2019 Nov 29;294(48):18387-18397. doi: 10.1074/jbc.RA119.009834. Epub 2019 Oct 9. J Biol Chem. 2019. PMID: 31597704 Free PMC article.
References
-
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. - PubMed
-
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

