Master transcription factors and mediator establish super-enhancers at key cell identity genes
- PMID: 23582322
- PMCID: PMC3653129
- DOI: 10.1016/j.cell.2013.03.035
Master transcription factors and mediator establish super-enhancers at key cell identity genes
Abstract
Master transcription factors Oct4, Sox2, and Nanog bind enhancer elements and recruit Mediator to activate much of the gene expression program of pluripotent embryonic stem cells (ESCs). We report here that the ESC master transcription factors form unusual enhancer domains at most genes that control the pluripotent state. These domains, which we call super-enhancers, consist of clusters of enhancers that are densely occupied by the master regulators and Mediator. Super-enhancers differ from typical enhancers in size, transcription factor density and content, ability to activate transcription, and sensitivity to perturbation. Reduced levels of Oct4 or Mediator cause preferential loss of expression of super-enhancer-associated genes relative to other genes, suggesting how changes in gene expression programs might be accomplished during development. In other more differentiated cells, super-enhancers containing cell-type-specific master transcription factors are also found at genes that define cell identity. Super-enhancers thus play key roles in the control of mammalian cell identity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
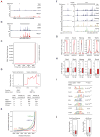
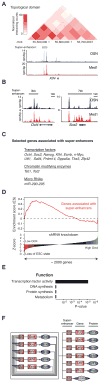
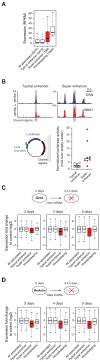


Comment in
-
Gene expression: Super enhancers.Nat Rev Genet. 2013 Jun;14(6):367. doi: 10.1038/nrg3496. Epub 2013 Apr 23. Nat Rev Genet. 2013. PMID: 23609410 No abstract available.
Similar articles
-
Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes.Mol Cell. 2019 Dec 5;76(5):753-766.e6. doi: 10.1016/j.molcel.2019.08.016. Epub 2019 Sep 25. Mol Cell. 2019. PMID: 31563432 Free PMC article.
-
Gene expression: Super enhancers.Nat Rev Genet. 2013 Jun;14(6):367. doi: 10.1038/nrg3496. Epub 2013 Apr 23. Nat Rev Genet. 2013. PMID: 23609410 No abstract available.
-
Super-enhancers in the control of cell identity and disease.Cell. 2013 Nov 7;155(4):934-47. doi: 10.1016/j.cell.2013.09.053. Epub 2013 Oct 10. Cell. 2013. PMID: 24119843 Free PMC article.
-
The molecular understanding of super-enhancer dysregulation in cancer.Nagoya J Med Sci. 2022 May;84(2):216-229. doi: 10.18999/nagjms.84.2.216. Nagoya J Med Sci. 2022. PMID: 35967935 Free PMC article. Review.
-
Enhancer RNAs: a missing regulatory layer in gene transcription.Sci China Life Sci. 2019 Jul;62(7):905-912. doi: 10.1007/s11427-017-9370-9. Epub 2018 Dec 26. Sci China Life Sci. 2019. PMID: 30593613 Review.
Cited by
-
The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation.Sci Immunol. 2021 Mar 12;6(57):eabc9801. doi: 10.1126/sciimmunol.abc9801. Sci Immunol. 2021. PMID: 33712472 Free PMC article.
-
RORγt expression in mature TH17 cells safeguards their lineage specification by inhibiting conversion to TH2 cells.Sci Adv. 2022 Aug 26;8(34):eabn7774. doi: 10.1126/sciadv.abn7774. Epub 2022 Aug 26. Sci Adv. 2022. PMID: 36026450 Free PMC article.
-
Tbx3 Controls Dppa3 Levels and Exit from Pluripotency toward Mesoderm.Stem Cell Reports. 2015 Jul 14;5(1):97-110. doi: 10.1016/j.stemcr.2015.05.009. Epub 2015 Jun 18. Stem Cell Reports. 2015. PMID: 26095607 Free PMC article.
-
Epigenomic profiling of glucocorticoid responses identifies cis-regulatory disruptions impacting steroid resistance in childhood acute lymphoblastic leukemia.Leukemia. 2022 Oct;36(10):2374-2383. doi: 10.1038/s41375-022-01685-z. Epub 2022 Aug 26. Leukemia. 2022. PMID: 36028659 Free PMC article.
-
Epigenetics of neural differentiation: Spotlight on enhancers.Front Cell Dev Biol. 2022 Oct 13;10:1001701. doi: 10.3389/fcell.2022.1001701. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313573 Free PMC article. Review.
References
-
- Abujarour R, Efe J, Ding S. Genome-wide gain-of-function screen identifies novel regulators of pluripotency. Stem Cells. 2010;28:1487–1497. - PubMed
-
- Alinikula J, Kohonen P, Nera KP, Lassila O. Concerted action of Helios and Ikaros controls the expression of the inositol 5-phosphatase SHIP. Eur J Immunol. 2010;40:2599–2607. - PubMed
-
- Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol. 2011;22:759–768. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

