A covalent protein-DNA 5'-product adduct is generated following AP lyase activity of human ALKBH1 (AlkB homologue 1)
- PMID: 23577621
- PMCID: PMC4126167
- DOI: 10.1042/BJ20121908
A covalent protein-DNA 5'-product adduct is generated following AP lyase activity of human ALKBH1 (AlkB homologue 1)
Abstract
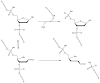
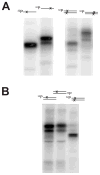
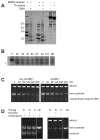
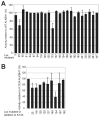
ALKBH1 (AlkB homologue 1) is a mammalian AlkB (2-oxoglutarate-dependent dioxygenase) homologue that possesses AP (abasic or apurinic/apyrimidinic) lyase activity. The AP lyase reaction is catalysed by imine formation with an active site lysine residue, and a covalent intermediate can be trapped in the presence of NaBH4. Surprisingly, ALKBH1 also forms a stable protein-DNA adduct in the absence of a reducing agent. Experiments with different substrates demonstrated that the protein covalently binds to the 5' DNA product, i.e. the fragment containing an α,β-unsaturated aldehyde. The N-terminal domain of ALKBH1 was identified as the main site of linkage with DNA. By contrast, mutagenesis studies suggest that the primary catalytic residue forming the imine linkage is Lys133, with Lys154 and other lysine residues in this region serving in opportunistic roles. These findings confirm the classification of ALKBH1 as an AP lyase, identify the primary and a secondary lysine residues involved in the lyase reaction, and demonstrate that the protein forms a covalent adduct with the 5' DNA product. We propose two plausible chemical mechanisms to account for the covalent attachment.
Figures



Similar articles
-
Characterization of human AlkB homolog 1 produced in mammalian cells and demonstration of mitochondrial dysfunction in ALKBH1-deficient cells.Biochem Biophys Res Commun. 2018 Jan 1;495(1):98-103. doi: 10.1016/j.bbrc.2017.10.158. Epub 2017 Oct 31. Biochem Biophys Res Commun. 2018. PMID: 29097205 Free PMC article.
-
Biochemical Characterization of AP Lyase and m6A Demethylase Activities of Human AlkB Homologue 1 (ALKBH1).Biochemistry. 2017 Apr 4;56(13):1899-1910. doi: 10.1021/acs.biochem.7b00060. Epub 2017 Mar 21. Biochemistry. 2017. PMID: 28290676 Free PMC article.
-
ALKBH1 is dispensable for abasic site cleavage during base excision repair and class switch recombination.PLoS One. 2013 Jun 25;8(6):e67403. doi: 10.1371/journal.pone.0067403. Print 2013. PLoS One. 2013. PMID: 23825659 Free PMC article.
-
Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases.Mol Biol Rep. 2021 May;48(5):4747-4756. doi: 10.1007/s11033-021-06421-x. Epub 2021 May 27. Mol Biol Rep. 2021. PMID: 34046849 Review.
-
[AP site recognition mechanism by the tryptophan residue in the vicinity of the catalytic center of AP endonucleases].Seikagaku. 2011 Jul;83(7):618-23. Seikagaku. 2011. PMID: 21866873 Review. Japanese. No abstract available.
Cited by
-
Spectroscopic and in vitro Investigations of Fe2+ /α-Ketoglutarate-Dependent Enzymes Involved in Nucleic Acid Repair and Modification.Chembiochem. 2022 Jun 3;23(11):e202100605. doi: 10.1002/cbic.202100605. Epub 2022 Feb 15. Chembiochem. 2022. PMID: 35040547 Free PMC article. Review.
-
Looking beneath the surface to determine what makes DNA damage deleterious.Curr Opin Chem Biol. 2014 Aug;21:48-55. doi: 10.1016/j.cbpa.2014.03.018. Epub 2014 Apr 22. Curr Opin Chem Biol. 2014. PMID: 24762292 Free PMC article. Review.
-
A DNA adenine demethylase impairs PRC2-mediated repression of genes marked by a specific chromatin signature.Genome Biol. 2023 Aug 30;24(1):198. doi: 10.1186/s13059-023-03042-4. Genome Biol. 2023. PMID: 37649077 Free PMC article.
-
Nucleic acid oxidation in DNA damage repair and epigenetics.Chem Rev. 2014 Apr 23;114(8):4602-20. doi: 10.1021/cr400432d. Epub 2014 Feb 28. Chem Rev. 2014. PMID: 24580634 Free PMC article. Review. No abstract available.
-
Multi-substrate selectivity based on key loops and non-homologous domains: new insight into ALKBH family.Cell Mol Life Sci. 2021 Jan;78(1):129-141. doi: 10.1007/s00018-020-03594-9. Epub 2020 Jul 8. Cell Mol Life Sci. 2021. PMID: 32642789 Free PMC article. Review.
References
-
- Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. - PubMed
-
- Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 1999;52:65–83. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

