Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity
- PMID: 23563690
- PMCID: PMC3652626
- DOI: 10.1038/ni.2570
Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity
Abstract
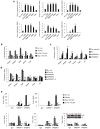
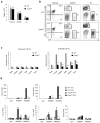
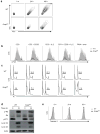
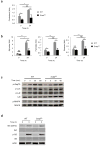
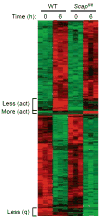
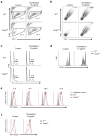
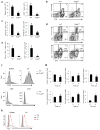
Newly activated CD8(+) T cells reprogram their metabolism to meet the extraordinary biosynthetic demands of clonal expansion; however, the signals that mediate metabolic reprogramming remain poorly defined. Here we demonstrate an essential role for sterol regulatory element-binding proteins (SREBPs) in the acquisition of effector-cell metabolism. Without SREBP signaling, CD8(+) T cells were unable to blast, which resulted in attenuated clonal expansion during viral infection. Mechanistic studies indicated that SREBPs were essential for meeting the heightened lipid requirements of membrane synthesis during blastogenesis. SREBPs were dispensable for homeostatic proliferation, which indicated a context-specific requirement for SREBPs in effector responses. Our studies provide insights into the molecular signals that underlie the metabolic reprogramming of CD8(+) T cells during the transition from quiescence to activation.
Figures

Similar articles
-
Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway.J Biol Chem. 2009 Apr 3;284(14):9237-46. doi: 10.1074/jbc.M808773200. Epub 2009 Feb 9. J Biol Chem. 2009. PMID: 19204002 Free PMC article.
-
Petroleum ether sub-fraction of rosemary extract improves hyperlipidemia and insulin resistance by inhibiting SREBPs.Chin J Nat Med. 2016 Oct;14(10):746-756. doi: 10.1016/S1875-5364(16)30089-9. Epub 2016 Oct 31. Chin J Nat Med. 2016. PMID: 28236404
-
Sterol regulatory element-binding proteins are transcriptional regulators of the thyroglobulin gene in thyroid cells.Biochim Biophys Acta. 2016 Aug;1859(8):994-1003. doi: 10.1016/j.bbagrm.2016.06.004. Epub 2016 Jun 16. Biochim Biophys Acta. 2016. PMID: 27321819
-
Effects of sterol regulatory element-binding protein (SREBP) in chickens.Lipids Health Dis. 2012 Feb 6;11:20. doi: 10.1186/1476-511X-11-20. Lipids Health Dis. 2012. PMID: 22309629 Free PMC article. Review.
-
Sterol regulatory element-binding protein family as global regulators of lipid synthetic genes in energy metabolism.Vitam Horm. 2002;65:167-94. doi: 10.1016/s0083-6729(02)65064-2. Vitam Horm. 2002. PMID: 12481547 Review.
Cited by
-
RhoA orchestrates glycolysis for TH2 cell differentiation and allergic airway inflammation.J Allergy Clin Immunol. 2016 Jan;137(1):231-245.e4. doi: 10.1016/j.jaci.2015.05.004. Epub 2015 Jun 19. J Allergy Clin Immunol. 2016. PMID: 26100081 Free PMC article.
-
TCR-Signaling Events in Cellular Metabolism and Specialization.Front Immunol. 2015 Jun 8;6:292. doi: 10.3389/fimmu.2015.00292. eCollection 2015. Front Immunol. 2015. PMID: 26106392 Free PMC article. Review.
-
Impact of cholesterol-pathways on breast cancer development, a metabolic landscape.J Cancer. 2021 May 19;12(14):4307-4321. doi: 10.7150/jca.54637. eCollection 2021. J Cancer. 2021. PMID: 34093831 Free PMC article. Review.
-
CD8+ T cell metabolism in infection and cancer.Nat Rev Immunol. 2021 Nov;21(11):718-738. doi: 10.1038/s41577-021-00537-8. Epub 2021 May 12. Nat Rev Immunol. 2021. PMID: 33981085 Free PMC article. Review.
-
Environmental and metabolic sensors that control T cell biology.Front Immunol. 2015 Mar 17;6:99. doi: 10.3389/fimmu.2015.00099. eCollection 2015. Front Immunol. 2015. PMID: 25852681 Free PMC article. Review.
References
-
- Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. - PubMed
-
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nature reviews. Immunology. 2005;5:844–852. - PubMed
-
- Rathmell JC, Elstrom RL, Cinalli RM, Thompson CB. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. European journal of immunology. 2003;33:2223–2232. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01 AI085043/AI/NIAID NIH HHS/United States
- T32-GM008469/GM/NIGMS NIH HHS/United States
- T32-CA009120-36/CA/NCI NIH HHS/United States
- T32 CA009120/CA/NCI NIH HHS/United States
- T32 GM008496/GM/NIGMS NIH HHS/United States
- AI082975/AI/NIAID NIH HHS/United States
- R01 AI093768/AI/NIAID NIH HHS/United States
- U01 AI082975/AI/NIAID NIH HHS/United States
- AI093768/AI/NIAID NIH HHS/United States
- AI085043/AI/NIAID NIH HHS/United States
- S10 RR026744/RR/NCRR NIH HHS/United States
- P01 HL090553/HL/NHLBI NIH HHS/United States
- R01 HL048044/HL/NHLBI NIH HHS/United States
- S10RR026744/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

