Nonenzymatic assembly of branched polyubiquitin chains for structural and biochemical studies
- PMID: 23557636
- PMCID: PMC3665622
- DOI: 10.1016/j.bmc.2013.02.052
Nonenzymatic assembly of branched polyubiquitin chains for structural and biochemical studies
Abstract
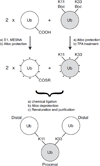
Polymeric chains of a small protein ubiquitin are involved in regulation of nearly all vital processes in eukaryotic cells. Elucidating the signaling properties of polyubiquitin requires the ability to make these chains in vitro. In recent years, chemical and chemical-biology tools have been developed that produce fully natural isopeptide-linked polyUb chains with no need for linkage-specific ubiquitin-conjugating enzymes. These methods produced unbranched chains (in which no more than one lysine per ubiquitin is conjugated to another ubiquitin). Here we report a nonenzymatic method for the assembly of fully natural isopeptide-linked branched polyubiquitin chains. This method is based on the use of mutually orthogonal removable protecting groups (e.g., Boc- and Alloc-) on lysines combined with an Ag-catalyzed condensation reaction between a C-terminal thioester on one ubiquitin and a specific ε-amine on another ubiquitin, and involves genetic incorporation of more than one Lys(Boc) at the desired linkage positions in the ubiquitin sequence. We demonstrate our method by making a fully natural branched tri-ubiquitin containing isopeptide linkages via Lys11 and Lys33, and a (15)N-enriched proximal ubiquitin, which enabled monomer-specific structural and dynamical studies by NMR. Furthermore, we assayed disassembly of branched and unbranched tri-ubiquitins as well as control di-ubiquitins by the yeast proteasome-associated deubiquitinase Ubp6. Our results show that Ubp6 can recognize and disassemble a branched polyubiquitin, wherein cleavage preferences for individual linkages are retained. Our spectroscopic and functional data suggest that, at least for the chains studied here, the isopeptide linkages are effectively independent of each other. Together with our method for nonenzymatic assembly of unbranched polyubiquitin, these developments now provide tools for making fully natural polyubiquitin chains of essentially any type of linkage and length.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Nonenzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme.J Am Chem Soc. 2011 Nov 9;133(44):17855-68. doi: 10.1021/ja207220g. Epub 2011 Oct 19. J Am Chem Soc. 2011. PMID: 21962295 Free PMC article.
-
Emerging roles for Lys11-linked polyubiquitin in cellular regulation.Trends Biochem Sci. 2011 Jul;36(7):355-63. doi: 10.1016/j.tibs.2011.04.004. Epub 2011 Jun 7. Trends Biochem Sci. 2011. PMID: 21641804 Review.
-
The Crystal Structure and Conformations of an Unbranched Mixed Tri-Ubiquitin Chain Containing K48 and K63 Linkages.J Mol Biol. 2017 Dec 8;429(24):3801-3813. doi: 10.1016/j.jmb.2017.10.027. Epub 2017 Oct 27. J Mol Biol. 2017. PMID: 29111344
-
Controlled synthesis of polyubiquitin chains.Methods Enzymol. 2005;399:21-36. doi: 10.1016/S0076-6879(05)99002-2. Methods Enzymol. 2005. PMID: 16338346
-
Reading the ubiquitin postal code.Curr Opin Struct Biol. 2011 Dec;21(6):792-801. doi: 10.1016/j.sbi.2011.09.009. Epub 2011 Oct 27. Curr Opin Struct Biol. 2011. PMID: 22036065 Review.
Cited by
-
Triubiquitin Probes for Identification of Reader and Eraser Proteins of Branched Polyubiquitin Chains.ACS Chem Biol. 2023 Apr 21;18(4):837-847. doi: 10.1021/acschembio.2c00898. Epub 2023 Mar 27. ACS Chem Biol. 2023. PMID: 36972492 Free PMC article.
-
Chemical biology approaches for studying posttranslational modifications.RNA Biol. 2018;15(4-5):427-440. doi: 10.1080/15476286.2017.1360468. Epub 2017 Sep 21. RNA Biol. 2018. PMID: 28901832 Free PMC article. Review.
-
Preparing to read the ubiquitin code: characterization of ubiquitin trimers by top-down mass spectrometry.J Mass Spectrom. 2016 Apr;51(4):315-21. doi: 10.1002/jms.3759. J Mass Spectrom. 2016. PMID: 27041663 Free PMC article.
-
Genetic Code Expansion Approaches to Decipher the Ubiquitin Code.Chem Rev. 2024 Oct 23;124(20):11544-11584. doi: 10.1021/acs.chemrev.4c00375. Epub 2024 Sep 23. Chem Rev. 2024. PMID: 39311880 Free PMC article. Review.
-
Chemical ubiquitination for decrypting a cellular code.Biochem J. 2016 May 15;473(10):1297-314. doi: 10.1042/BJ20151195. Biochem J. 2016. PMID: 27208213 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

