Regulation of stress granules and P-bodies during RNA virus infection
- PMID: 23554219
- PMCID: PMC3652661
- DOI: 10.1002/wrna.1162
Regulation of stress granules and P-bodies during RNA virus infection
Abstract
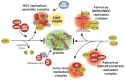
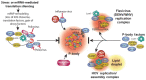
RNA granules are structures within cells that play major roles in gene expression and homeostasis. Two principle kinds of RNA granules are conserved from yeast to mammals: stress granules (SGs), which contain stalled translation initiation complexes, and processing bodies (P-bodies, PBs), which are enriched with factors involved in RNA turnover. Since RNA granules are associated with silenced transcripts, viruses subvert RNA granule function for replicative advantages. This review, focusing on RNA viruses, discusses mechanisms that manipulate stress granules and P-bodies to promote synthesis of viral proteins. Three main themes have emerged for how viruses manipulate RNA granules; (1) cleavage of key host factors, (2) control of protein kinase R (PKR) activation, and (3) redirecting RNA granule components for new or parallel roles in viral reproduction, at the same time disrupting RNA granules. Viruses utilize one or more of these routes to achieve robust and productive infection.
Copyright © 2013 John Wiley & Sons, Ltd.
Figures
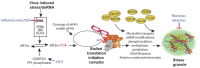
Similar articles
-
Diversion of stress granules and P-bodies during viral infection.Virology. 2013 Feb 20;436(2):255-67. doi: 10.1016/j.virol.2012.11.017. Epub 2013 Jan 3. Virology. 2013. PMID: 23290869 Free PMC article. Review.
-
Cytoplasmic RNA Granules and Viral Infection.Annu Rev Virol. 2014 Nov;1(1):147-70. doi: 10.1146/annurev-virology-031413-085505. Annu Rev Virol. 2014. PMID: 26958719 Free PMC article.
-
Chandipura Virus Forms Cytoplasmic Inclusion Bodies through Phase Separation and Proviral Association of Cellular Protein Kinase R and Stress Granule Protein TIA-1.Viruses. 2024 Jun 26;16(7):1027. doi: 10.3390/v16071027. Viruses. 2024. PMID: 39066190 Free PMC article.
-
Mouse Norovirus Infection Arrests Host Cell Translation Uncoupled from the Stress Granule-PKR-eIF2α Axis.mBio. 2019 Jun 18;10(3):e00960-19. doi: 10.1128/mBio.00960-19. mBio. 2019. PMID: 31213553 Free PMC article.
-
Who Regulates Whom? An Overview of RNA Granules and Viral Infections.Viruses. 2016 Jun 28;8(7):180. doi: 10.3390/v8070180. Viruses. 2016. PMID: 27367717 Free PMC article. Review.
Cited by
-
Post-transcriptional coordination of immunological responses by RNA-binding proteins.Nat Immunol. 2014 Jun;15(6):492-502. doi: 10.1038/ni.2884. Nat Immunol. 2014. PMID: 24840980 Review.
-
CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication.Elife. 2020 Mar 24;9:e53753. doi: 10.7554/eLife.53753. Elife. 2020. PMID: 32207684 Free PMC article.
-
Induction of stress granules by interferon and down-regulation by the cellular RNA adenosine deaminase ADAR1.Virology. 2014 Apr;454-455:299-310. doi: 10.1016/j.virol.2014.02.025. Epub 2014 Mar 21. Virology. 2014. PMID: 24725957 Free PMC article.
-
Genomic profiling of collaborative cross founder mice infected with respiratory viruses reveals novel transcripts and infection-related strain-specific gene and isoform expression.G3 (Bethesda). 2014 Jun 5;4(8):1429-44. doi: 10.1534/g3.114.011759. G3 (Bethesda). 2014. PMID: 24902603 Free PMC article.
-
Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly.J Virol. 2017 Feb 28;91(6):e02315-16. doi: 10.1128/JVI.02315-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053097 Free PMC article.
References
-
- Chang T‐C, Yamashita A, Chen C‐YA, Yamashita Y, Zhu W, Durdan S, Kahvejian A, Sonenberg N, Shyu A‐B. UNR, a new partner of poly(A)‐binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c‐fos major coding‐region determinant. Genes Dev 2004, 18:2010–2023. - PMC - PubMed
-
- Caudron‐Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev 2012, 22:179–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

