The genomic and transcriptomic landscape of a HeLa cell line
- PMID: 23550136
- PMCID: PMC3737162
- DOI: 10.1534/g3.113.005777
The genomic and transcriptomic landscape of a HeLa cell line
Abstract
HeLa is the most widely used model cell line for studying human cellular and molecular biology. To date, no genomic reference for this cell line has been released, and experiments have relied on the human reference genome. Effective design and interpretation of molecular genetic studies performed using HeLa cells require accurate genomic information. Here we present a detailed genomic and transcriptomic characterization of a HeLa cell line. We performed DNA and RNA sequencing of a HeLa Kyoto cell line and analyzed its mutational portfolio and gene expression profile. Segmentation of the genome according to copy number revealed a remarkably high level of aneuploidy and numerous large structural variants at unprecedented resolution. Some of the extensive genomic rearrangements are indicative of catastrophic chromosome shattering, known as chromothripsis. Our analysis of the HeLa gene expression profile revealed that several pathways, including cell cycle and DNA repair, exhibit significantly different expression patterns from those in normal human tissues. Our results provide the first detailed account of genomic variants in the HeLa genome, yielding insight into their impact on gene expression and cellular function as well as their origins. This study underscores the importance of accounting for the strikingly aberrant characteristics of HeLa cells when designing and interpreting experiments, and has implications for the use of HeLa as a model of human biology.
Keywords: HeLa cell line; genomics; resource; transcriptomics; variation.
Figures
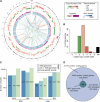
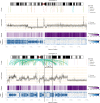

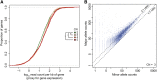
Comment in
-
The fractured genome of HeLa cells.Genome Biol. 2013 Apr 17;14(4):111. doi: 10.1186/gb-2013-14-4-111. Genome Biol. 2013. PMID: 23594443 Free PMC article.
Similar articles
-
The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line.Nature. 2013 Aug 8;500(7461):207-11. doi: 10.1038/nature12064. Nature. 2013. PMID: 23925245 Free PMC article.
-
High variability of genomic instability and gene expression profiling in different HeLa clones.Sci Rep. 2015 Oct 20;5:15377. doi: 10.1038/srep15377. Sci Rep. 2015. PMID: 26483214 Free PMC article.
-
HeLa-CCL2 cell heterogeneity studied by single-cell DNA and RNA sequencing.PLoS One. 2019 Dec 2;14(12):e0225466. doi: 10.1371/journal.pone.0225466. eCollection 2019. PLoS One. 2019. PMID: 31790455 Free PMC article.
-
Genomics and the Immune Landscape of Osteosarcoma.Adv Exp Med Biol. 2020;1258:21-36. doi: 10.1007/978-3-030-43085-6_2. Adv Exp Med Biol. 2020. PMID: 32767232 Review.
-
Understanding genomic alterations in cancer genomes using an integrative network approach.Cancer Lett. 2013 Nov 1;340(2):261-9. doi: 10.1016/j.canlet.2012.11.050. Epub 2012 Dec 22. Cancer Lett. 2013. PMID: 23266571 Review.
Cited by
-
Editorial: HeLa cell sequela.G3 (Bethesda). 2013 Jun 21;3(6):927. doi: 10.1534/g3.113.006304. G3 (Bethesda). 2013. PMID: 23576522 Free PMC article. No abstract available.
-
Peak-valley-peak pattern of histone modifications delineates active regulatory elements and their directionality.Nucleic Acids Res. 2016 May 19;44(9):4037-51. doi: 10.1093/nar/gkw250. Epub 2016 Apr 19. Nucleic Acids Res. 2016. PMID: 27095194 Free PMC article.
-
Intranuclear dynamics of the Nup107-160 complex.Mol Biol Cell. 2015 Jun 15;26(12):2343-56. doi: 10.1091/mbc.E15-02-0060. Epub 2015 Apr 22. Mol Biol Cell. 2015. PMID: 25904327 Free PMC article.
-
Major changes of cell function and toxicant sensitivity in cultured cells undergoing mild, quasi-natural genetic drift.Arch Toxicol. 2018 Dec;92(12):3487-3503. doi: 10.1007/s00204-018-2326-5. Epub 2018 Oct 8. Arch Toxicol. 2018. PMID: 30298209 Free PMC article.
-
Global detection of human variants and isoforms by deep proteome sequencing.Nat Biotechnol. 2023 Dec;41(12):1776-1786. doi: 10.1038/s41587-023-01714-x. Epub 2023 Mar 23. Nat Biotechnol. 2023. PMID: 36959352 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
