Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen
- PMID: 23545501
- PMCID: PMC3748285
- DOI: 10.1016/j.celrep.2013.03.002
Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen
Abstract
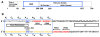
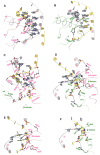
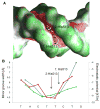
The DNA tumor virus Simian virus 40 (SV40) is a model system for studying eukaryotic replication. SV40 large tumor antigen (LTag) is the initiator/helicase that is essential for genome replication. LTag recognizes and assembles at the viral replication origin. We determined the structure of two multidomain LTag subunits bound to origin DNA. The structure reveals that the origin binding domains (OBDs) and Zn and AAA+ domains are involved in origin recognition and assembly. Notably, the OBDs recognize the origin in an unexpected manner. The histidine residues of the AAA+ domains insert into a narrow minor groove region with enhanced negative electrostatic potential. Computational analysis indicates that this region is intrinsically narrow, demonstrating the role of DNA shape readout in origin recognition. Our results provide important insights into the assembly of the LTag initiator/helicase at the replication origin and suggest that histidine contacts with the minor groove serve as a mechanism of DNA shape readout.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
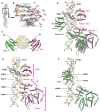
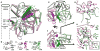
Similar articles
-
Conformational rearrangements of SV40 large T antigen during early replication events.J Mol Biol. 2010 Apr 16;397(5):1276-86. doi: 10.1016/j.jmb.2010.02.042. Epub 2010 Feb 26. J Mol Biol. 2010. PMID: 20219473 Free PMC article.
-
A model for polyomavirus helicase activity derived in part from the AlphaFold2 structure of SV40 T-antigen.J Virol. 2024 Oct 22;98(10):e0111924. doi: 10.1128/jvi.01119-24. Epub 2024 Sep 23. J Virol. 2024. PMID: 39311578
-
The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA.Elife. 2016 Dec 6;5:e18129. doi: 10.7554/eLife.18129. Elife. 2016. Retraction in: Elife. 2019 Apr 03;8:e46910. doi: 10.7554/eLife.46910 PMID: 27921994 Free PMC article. Retracted.
-
The initiation of simian virus 40 DNA replication in vitro.Crit Rev Biochem Mol Biol. 1997;32(6):503-68. doi: 10.3109/10409239709082001. Crit Rev Biochem Mol Biol. 1997. PMID: 9444478 Review.
-
The MCM complex: (just) a replicative helicase?Biochem Soc Trans. 2008 Feb;36(Pt 1):136-40. doi: 10.1042/BST0360136. Biochem Soc Trans. 2008. PMID: 18208401 Review.
Cited by
-
Quantitative modeling of transcription factor binding specificities using DNA shape.Proc Natl Acad Sci U S A. 2015 Apr 14;112(15):4654-9. doi: 10.1073/pnas.1422023112. Epub 2015 Mar 9. Proc Natl Acad Sci U S A. 2015. PMID: 25775564 Free PMC article.
-
Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding.Nucleic Acids Res. 2017 Dec 15;45(22):12877-12887. doi: 10.1093/nar/gkx1145. Nucleic Acids Res. 2017. PMID: 29165643 Free PMC article.
-
Genome-wide prediction of minor-groove electrostatic potential enables biophysical modeling of protein-DNA binding.Nucleic Acids Res. 2017 Dec 1;45(21):12565-12576. doi: 10.1093/nar/gkx915. Nucleic Acids Res. 2017. PMID: 29040720 Free PMC article.
-
Probing the role of the protonation state of a minor groove-linker histidine in Exd-Hox-DNA binding.Biophys J. 2024 Jan 16;123(2):248-259. doi: 10.1016/j.bpj.2023.12.013. Epub 2023 Dec 21. Biophys J. 2024. PMID: 38130056 Free PMC article.
-
Deconvolving the recognition of DNA shape from sequence.Cell. 2015 Apr 9;161(2):307-18. doi: 10.1016/j.cell.2015.02.008. Epub 2015 Apr 2. Cell. 2015. PMID: 25843630 Free PMC article.
References
-
- Borowiec JA, Dean FB, Bullock PA, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

