Torque measurement at the single-molecule level
- PMID: 23541162
- PMCID: PMC5515228
- DOI: 10.1146/annurev-biophys-083012-130412
Torque measurement at the single-molecule level
Abstract
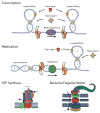
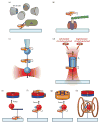
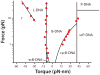
Methods for exerting and measuring forces on single molecules have revolutionized the study of the physics of biology. However, it is often the case that biological processes involve rotation or torque generation, and these parameters have been more difficult to access experimentally. Recent advances in the single-molecule field have led to the development of techniques that add the capability of torque measurement. By combining force, displacement, torque, and rotational data, a more comprehensive description of the mechanics of a biomolecule can be achieved. In this review, we highlight a number of biological processes for which torque plays a key mechanical role. We describe the various techniques that have been developed to directly probe the torque experienced by a single molecule, and detail a variety of measurements made to date using these new technologies. We conclude by discussing a number of open questions and propose systems of study that would be well suited for analysis with torsional measurement techniques.
Figures

Similar articles
-
Angular Optical Trapping to Directly Measure DNA Torsional Mechanics.Methods Mol Biol. 2022;2478:37-73. doi: 10.1007/978-1-0716-2229-2_4. Methods Mol Biol. 2022. PMID: 36063318
-
Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments.Nat Methods. 2010 Dec;7(12):977-80. doi: 10.1038/nmeth.1520. Epub 2010 Oct 17. Nat Methods. 2010. PMID: 20953173
-
Combined Force-Torque Spectroscopy of Proteins by Means of Multiscale Molecular Simulation.Biophys J. 2020 Dec 1;119(11):2240-2250. doi: 10.1016/j.bpj.2020.09.039. Epub 2020 Oct 27. Biophys J. 2020. PMID: 33121942 Free PMC article.
-
Probing the mechanical properties, conformational changes, and interactions of nucleic acids with magnetic tweezers.J Struct Biol. 2017 Jan;197(1):26-36. doi: 10.1016/j.jsb.2016.06.022. Epub 2016 Jun 29. J Struct Biol. 2017. PMID: 27368129 Review.
-
Rotary protein motors.Trends Cell Biol. 2003 Mar;13(3):114-21. doi: 10.1016/s0962-8924(03)00004-7. Trends Cell Biol. 2003. PMID: 12628343 Review.
Cited by
-
DNA Y structure: a versatile, multidimensional single molecule assay.Nano Lett. 2014 Nov 12;14(11):6475-80. doi: 10.1021/nl503009d. Epub 2014 Oct 14. Nano Lett. 2014. PMID: 25291441 Free PMC article.
-
Dynamics of the Buckling Transition in Double-Stranded DNA and RNA.Biophys J. 2020 Apr 7;118(7):1690-1701. doi: 10.1016/j.bpj.2020.01.049. Epub 2020 Feb 29. Biophys J. 2020. PMID: 32367807 Free PMC article.
-
The dynamic interplay between DNA topoisomerases and DNA topology.Biophys Rev. 2016 Nov;8(Suppl 1):101-111. doi: 10.1007/s12551-016-0240-8. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28510219 Free PMC article. Review.
-
Blind predictions of DNA and RNA tweezers experiments with force and torque.PLoS Comput Biol. 2014 Aug 7;10(8):e1003756. doi: 10.1371/journal.pcbi.1003756. eCollection 2014 Aug. PLoS Comput Biol. 2014. PMID: 25102226 Free PMC article.
-
Understanding the Relative Flexibility of RNA and DNA Duplexes: Stretching and Twist-Stretch Coupling.Biophys J. 2017 Mar 28;112(6):1094-1104. doi: 10.1016/j.bpj.2017.02.022. Biophys J. 2017. PMID: 28355538 Free PMC article.
References
-
- Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. - PubMed
-
- Ashkin A, Dziedzic JM, Yamane T. Optical trapping and manipulation of single cells using infrared-laser beams. Nature. 1987;330:769–71. - PubMed
-
- Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand JF, et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13:444–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

