YY1 controls Igκ repertoire and B-cell development, and localizes with condensin on the Igκ locus
- PMID: 23531880
- PMCID: PMC3630362
- DOI: 10.1038/emboj.2013.66
YY1 controls Igκ repertoire and B-cell development, and localizes with condensin on the Igκ locus
Abstract
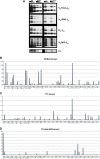
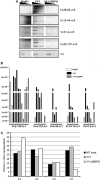
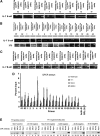
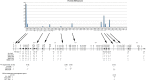
Conditional knock-out (KO) of Polycomb Group (PcG) protein YY1 results in pro-B cell arrest and reduced immunoglobulin locus contraction needed for distal variable gene rearrangement. The mechanisms that control these crucial functions are unknown. We deleted the 25 amino-acid YY1 REPO domain necessary for YY1 PcG function, and used this mutant (YY1ΔREPO), to transduce bone marrow from YY1 conditional KO mice. While wild-type YY1 rescued B-cell development, YY1ΔREPO failed to rescue the B-cell lineage yielding reduced numbers of B lineage cells. Although the IgH rearrangement pattern was normal, there was a selective impact at the Igκ locus that showed a dramatic skewing of the expressed Igκ repertoire. We found that the REPO domain interacts with proteins from the condensin and cohesin complexes, and that YY1, EZH2 and condensin proteins co-localize at numerous sites across the Ig kappa locus. Knock-down of a condensin subunit protein or YY1 reduced rearrangement of Igκ Vκ genes suggesting a direct role for YY1-condensin complexes in Igκ locus structure and rearrangement.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
YY1 binds to the E3' enhancer and inhibits the expression of the immunoglobulin κ gene via epigenetic modifications.Immunology. 2018 Dec;155(4):491-498. doi: 10.1111/imm.12990. Epub 2018 Aug 29. Immunology. 2018. PMID: 30098214 Free PMC article.
-
Increased expression of PcG protein YY1 negatively regulates B cell development while allowing accumulation of myeloid cells and LT-HSC cells.PLoS One. 2012;7(1):e30656. doi: 10.1371/journal.pone.0030656. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22292011 Free PMC article.
-
Epigenetic Enhancer Marks and Transcription Factor Binding Influence Vκ Gene Rearrangement in Pre-B Cells and Pro-B Cells.Front Immunol. 2018 Sep 13;9:2074. doi: 10.3389/fimmu.2018.02074. eCollection 2018. Front Immunol. 2018. PMID: 30271408 Free PMC article.
-
Function of YY1 in Long-Distance DNA Interactions.Front Immunol. 2014 Feb 10;5:45. doi: 10.3389/fimmu.2014.00045. eCollection 2014. Front Immunol. 2014. PMID: 24575094 Free PMC article. Review.
-
Dynamic Control of Long-Range Genomic Interactions at the Immunoglobulin κ Light-Chain Locus.Adv Immunol. 2015;128:183-271. doi: 10.1016/bs.ai.2015.07.004. Epub 2015 Aug 14. Adv Immunol. 2015. PMID: 26477368 Review.
Cited by
-
The Myc-associated zinc finger protein (MAZ) works together with CTCF to control cohesin positioning and genome organization.Proc Natl Acad Sci U S A. 2021 Feb 16;118(7):e2023127118. doi: 10.1073/pnas.2023127118. Proc Natl Acad Sci U S A. 2021. PMID: 33558242 Free PMC article.
-
The Two Sides of YY1 in Cancer: A Friend and a Foe.Front Oncol. 2019 Nov 20;9:1230. doi: 10.3389/fonc.2019.01230. eCollection 2019. Front Oncol. 2019. PMID: 31824839 Free PMC article. Review.
-
Comparing and Contrasting the Effects of Drosophila Condensin II Subunit dCAP-D3 Overexpression and Depletion in Vivo.Genetics. 2018 Oct;210(2):531-546. doi: 10.1534/genetics.118.301344. Epub 2018 Aug 1. Genetics. 2018. PMID: 30068527 Free PMC article.
-
Targeting Transcription Factor YY1 for Cancer Treatment: Current Strategies and Future Directions.Cancers (Basel). 2023 Jul 5;15(13):3506. doi: 10.3390/cancers15133506. Cancers (Basel). 2023. PMID: 37444616 Free PMC article. Review.
-
A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation.Genes Dev. 2015 Aug 15;29(16):1683-95. doi: 10.1101/gad.263871.115. Genes Dev. 2015. PMID: 26302788 Free PMC article.
References
-
- Akasaka T, Tsuji K-i, Kawahira H, Kanno M, Harigaya K-i, Hu L, Ebihara Y, Nakahata T, Tetsu O, Taniguchi M, Koseki H (1997) The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity 7: 135–146 - PubMed
-
- Alkema MJ, van der Lugt NMT, Bobeldijk RC, Berns A, van Lohuizen M (1995) Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature 374: 724–727 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

