Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome
- PMID: 23531545
- PMCID: PMC3643606
- DOI: 10.1093/nar/gkt203
Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome
Abstract
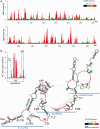
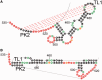
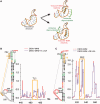
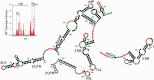
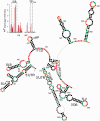
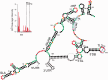
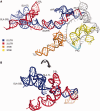
The Dengue virus (DENV) genome contains multiple cis-acting elements required for translation and replication. Previous studies indicated that a 719-nt subgenomic minigenome (DENV-MINI) is an efficient template for translation and (-) strand RNA synthesis in vitro. We performed a detailed structural analysis of DENV-MINI RNA, combining chemical acylation techniques, Pb(2+) ion-induced hydrolysis and site-directed mutagenesis. Our results highlight protein-independent 5'-3' terminal interactions involving hybridization between recognized cis-acting motifs. Probing analyses identified tandem dumbbell structures (DBs) within the 3' terminus spaced by single-stranded regions, internal loops and hairpins with embedded GNRA-like motifs. Analysis of conserved motifs and top loops (TLs) of these dumbbells, and their proposed interactions with downstream pseudoknot (PK) regions, predicted an H-type pseudoknot involving TL1 of the 5' DB and the complementary region, PK2. As disrupting the TL1/PK2 interaction, via 'flipping' mutations of PK2, previously attenuated DENV replication, this pseudoknot may participate in regulation of RNA synthesis. Computer modeling implied that this motif might function as autonomous structural/regulatory element. In addition, our studies targeting elements of the 3' DB and its complementary region PK1 indicated that communication between 5'-3' terminal regions strongly depends on structure and sequence composition of the 5' cyclization region.
Figures
Similar articles
-
Identification of cis-acting elements in the 3'-untranslated region of the dengue virus type 2 RNA that modulate translation and replication.J Biol Chem. 2011 Jun 24;286(25):22521-34. doi: 10.1074/jbc.M111.234302. Epub 2011 Apr 22. J Biol Chem. 2011. PMID: 21515677 Free PMC article.
-
The 5'-terminal region of the Aichi virus genome encodes cis-acting replication elements required for positive- and negative-strand RNA synthesis.J Virol. 2005 Jun;79(11):6918-31. doi: 10.1128/JVI.79.11.6918-6931.2005. J Virol. 2005. PMID: 15890931 Free PMC article.
-
Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization.J Virol. 2013 Jun;87(12):6804-18. doi: 10.1128/JVI.00243-13. Epub 2013 Apr 10. J Virol. 2013. PMID: 23576500 Free PMC article.
-
Structure and function of cis-acting RNA elements of flavivirus.Rev Med Virol. 2020 Jan;30(1):e2092. doi: 10.1002/rmv.2092. Epub 2019 Nov 28. Rev Med Virol. 2020. PMID: 31777997 Review.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
Cited by
-
Functions of the 3' and 5' genome RNA regions of members of the genus Flavivirus.Virus Res. 2015 Aug 3;206:108-19. doi: 10.1016/j.virusres.2015.02.006. Epub 2015 Feb 13. Virus Res. 2015. PMID: 25683510 Free PMC article. Review.
-
Flavors of Flaviviral RNA Structure: towards an Integrated View of RNA Function from Translation through Encapsidation.Bioessays. 2019 Aug;41(8):e1900003. doi: 10.1002/bies.201900003. Epub 2019 Jun 18. Bioessays. 2019. PMID: 31210384 Free PMC article. Review.
-
Structures and Functions of the 3' Untranslated Regions of Positive-Sense Single-Stranded RNA Viruses Infecting Humans and Animals.Front Cell Infect Microbiol. 2020 Aug 27;10:453. doi: 10.3389/fcimb.2020.00453. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32974223 Free PMC article. Review.
-
Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication.J Virol. 2015 Mar;89(6):3430-7. doi: 10.1128/JVI.02677-14. Epub 2015 Jan 14. J Virol. 2015. PMID: 25589642 Free PMC article.
-
What Does the Future Hold for Yellow Fever Virus? (II).Genes (Basel). 2018 Aug 21;9(9):425. doi: 10.3390/genes9090425. Genes (Basel). 2018. PMID: 30134625 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

