Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission
- PMID: 23530241
- PMCID: PMC3625255
- DOI: 10.1073/pnas.1300855110
Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission
Abstract
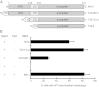
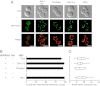
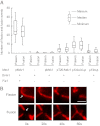
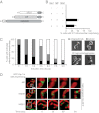
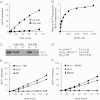
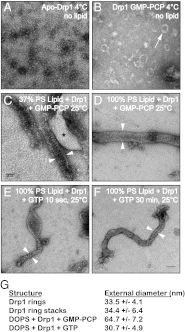
Mitochondrial fission is mediated by the dynamin-related GTPases Dnm1/Drp1 (yeast/mammals), which form spirals around constricted sites on mitochondria. Additional membrane-associated adaptor proteins (Fis1, Mdv1, Mff, and MiDs) are required to recruit these GTPases from the cytoplasm to the mitochondrial surface. Whether these adaptors participate in both GTPase recruitment and membrane scission is not known. Here we use a yeast strain lacking all fission proteins to identify the minimal combinations of GTPases and adaptors sufficient for mitochondrial fission. Although Fis1 is dispensable for fission, membrane-anchored Mdv1, Mff, or MiDs paired individually with their respective GTPases are sufficient to divide mitochondria. In addition to their role in Drp1 membrane recruitment, MiDs coassemble with Drp1 in vitro. The resulting heteropolymer adopts a dramatically different structure with a narrower diameter than Drp1 homopolymers assembled in isolation. This result demonstrates that an adaptor protein alters the architecture of a mitochondrial dynamin GTPase polymer in a manner that could facilitate membrane constriction and severing activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes.J Cell Biol. 2010 Dec 13;191(6):1127-39. doi: 10.1083/jcb.201005046. J Cell Biol. 2010. PMID: 21149566 Free PMC article.
-
Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission.Nat Struct Mol Biol. 2011 Jan;18(1):20-6. doi: 10.1038/nsmb.1949. Epub 2010 Dec 19. Nat Struct Mol Biol. 2011. PMID: 21170049 Free PMC article.
-
The mitochondrial fission adaptors Caf4 and Mdv1 are not functionally equivalent.PLoS One. 2012;7(12):e53523. doi: 10.1371/journal.pone.0053523. Epub 2012 Dec 31. PLoS One. 2012. PMID: 23300936 Free PMC article.
-
The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy.Int J Mol Sci. 2023 Mar 17;24(6):5785. doi: 10.3390/ijms24065785. Int J Mol Sci. 2023. PMID: 36982862 Free PMC article. Review.
-
The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: implications for human disease.Clin Sci (Lond). 2016 Nov 1;130(21):1861-74. doi: 10.1042/CS20160030. Clin Sci (Lond). 2016. PMID: 27660309 Review.
Cited by
-
The membrane remodeling protein Pex11p activates the GTPase Dnm1p during peroxisomal fission.Proc Natl Acad Sci U S A. 2015 May 19;112(20):6377-82. doi: 10.1073/pnas.1418736112. Epub 2015 May 4. Proc Natl Acad Sci U S A. 2015. PMID: 25941407 Free PMC article.
-
The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction.J Biol Chem. 2015 May 1;290(18):11692-703. doi: 10.1074/jbc.M114.610881. Epub 2015 Mar 13. J Biol Chem. 2015. PMID: 25770210 Free PMC article.
-
Molecular Basis of Mitochondrial and Peroxisomal Division Machineries.Int J Mol Sci. 2020 Jul 30;21(15):5452. doi: 10.3390/ijms21155452. Int J Mol Sci. 2020. PMID: 32751702 Free PMC article. Review.
-
The Role of Mitochondrial Dynamics and Mitotic Fission in Regulating the Cell Cycle in Cancer and Pulmonary Arterial Hypertension: Implications for Dynamin-Related Protein 1 and Mitofusin2 in Hyperproliferative Diseases.Cells. 2023 Jul 20;12(14):1897. doi: 10.3390/cells12141897. Cells. 2023. PMID: 37508561 Free PMC article. Review.
-
The mitochondrial fission receptor MiD51 requires ADP as a cofactor.Structure. 2014 Mar 4;22(3):367-77. doi: 10.1016/j.str.2014.01.001. Epub 2014 Feb 6. Structure. 2014. PMID: 24508339 Free PMC article.
References
-
- Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5(2):133–147. - PubMed
-
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282(15):11521–11529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

