Insight into the evolution of the histidine triad protein (HTP) family in Streptococcus
- PMID: 23527301
- PMCID: PMC3603884
- DOI: 10.1371/journal.pone.0060116
Insight into the evolution of the histidine triad protein (HTP) family in Streptococcus
Abstract
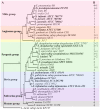
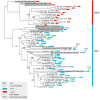
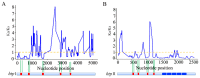
The Histidine Triad Proteins (HTPs), also known as Pht proteins in Streptococcus pneumoniae, constitute a family of surface-exposed proteins that exist in many pathogenic streptococcal species. Although many studies have revealed the importance of HTPs in streptococcal physiology and pathogenicity, little is known about their origin and evolution. In this study, after identifying all htp homologs from 105 streptococcal genomes representing 38 different species/subspecies, we analyzed their domain structures, positions in genome, and most importantly, their evolutionary histories. By further projecting this information onto the streptococcal phylogeny, we made several major findings. First, htp genes originated earlier than the Streptococcus genus and gene-loss events have occurred among three streptococcal groups, resulting in the absence of the htp gene in the Bovis, Mutans and Salivarius groups. Second, the copy number of htp genes in other groups of Streptococcus is variable, ranging from one to four functional copies. Third, both phylogenetic evidence and domain structure analyses support the division of two htp subfamilies, designated as htp I and htp II. Although present mainly in the pyogenic group and in Streptococcus suis, htp II members are distinct from htp I due to the presence of an additional leucine-rich-repeat domain at the C-terminus. Finally, htp genes exhibit a faster nucleotide substitution rate than do housekeeping genes. Specifically, the regions outside the HTP domains are under strong positive selection. This distinct evolutionary pattern likely helped Streptococcus to easily escape from recognition by host immunity.
Conflict of interest statement
Figures
Similar articles
-
The type II histidine triad protein HtpsC is a novel adhesion with the involvement of Streptococcus suis virulence.Virulence. 2015;6(6):631-41. doi: 10.1080/21505594.2015.1056971. Virulence. 2015. PMID: 26151575 Free PMC article.
-
Occurrence and evolution of the paralogous zinc metalloproteases IgA1 protease, ZmpB, ZmpC, and ZmpD in Streptococcus pneumoniae and related commensal species.mBio. 2012 Oct 2;3(5):e00303-12. doi: 10.1128/mBio.00303-12. Print 2012. mBio. 2012. PMID: 23033471 Free PMC article.
-
Molecular phylogeny and a taxonomic proposal for the genus Streptococcus.Genet Mol Res. 2015 Sep 21;14(3):10905-18. doi: 10.4238/2015.September.21.1. Genet Mol Res. 2015. PMID: 26400318
-
Genomics of Streptococcus salivarius, a major human commensal.Infect Genet Evol. 2015 Jul;33:381-92. doi: 10.1016/j.meegid.2014.10.001. Epub 2014 Oct 13. Infect Genet Evol. 2015. PMID: 25311532 Review.
-
Independent evolution of competence regulatory cascades in streptococci?Trends Microbiol. 2006 Aug;14(8):339-45. doi: 10.1016/j.tim.2006.06.007. Epub 2006 Jul 3. Trends Microbiol. 2006. PMID: 16820295 Review.
Cited by
-
Identification and validation of novel genes related to immune microenvironment in polycystic ovary syndrome.Medicine (Baltimore). 2024 Oct 25;103(43):e40229. doi: 10.1097/MD.0000000000040229. Medicine (Baltimore). 2024. PMID: 39470566 Free PMC article.
-
Zn2+ Uptake in Streptococcus pyogenes: Characterization of adcA and lmb Null Mutants.PLoS One. 2016 Mar 31;11(3):e0152835. doi: 10.1371/journal.pone.0152835. eCollection 2016. PLoS One. 2016. PMID: 27031880 Free PMC article.
-
Role of Pht proteins in attachment of Streptococcus pneumoniae to respiratory epithelial cells.Infect Immun. 2014 Apr;82(4):1683-91. doi: 10.1128/IAI.00699-13. Epub 2014 Feb 3. Infect Immun. 2014. PMID: 24491577 Free PMC article.
-
Overlapping functionality of the Pht proteins in zinc homeostasis of Streptococcus pneumoniae.Infect Immun. 2014 Oct;82(10):4315-24. doi: 10.1128/IAI.02155-14. Epub 2014 Jul 28. Infect Immun. 2014. PMID: 25069983 Free PMC article.
-
The findings of glucosyltransferase enzymes derived from oral streptococci.Jpn Dent Sci Rev. 2022 Nov;58:328-335. doi: 10.1016/j.jdsr.2022.10.003. Epub 2022 Oct 28. Jpn Dent Sci Rev. 2022. PMID: 36340584 Free PMC article. Review.
References
-
- Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T (1995) Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 45: 406–408. - PubMed
-
- Jarva H, Jokiranta TS, Wurzner R, Meri S (2003) Complement resistance mechanisms of streptococci. Mol Immunol 40: 95–107. - PubMed
-
- Soriani M, Telford JL (2010) Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol 5: 735–747. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

