Widespread and extensive lengthening of 3' UTRs in the mammalian brain
- PMID: 23520388
- PMCID: PMC3638137
- DOI: 10.1101/gr.146886.112
Widespread and extensive lengthening of 3' UTRs in the mammalian brain
Abstract
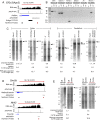
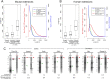
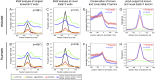
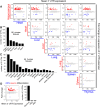
Remarkable advances in techniques for gene expression profiling have radically changed our knowledge of the transcriptome. Recently, the mammalian brain was reported to express many long intergenic noncoding (lincRNAs) from loci downstream from protein-coding genes. Our experimental tests failed to validate specific accumulation of lincRNA transcripts, and instead revealed strongly distal 3' UTRs generated by alternative cleavage and polyadenylation (APA). With this perspective in mind, we analyzed deep mammalian RNA-seq data using conservative criteria, and identified 2035 mouse and 1847 human genes that utilize substantially distal novel 3' UTRs. Each of these extends at least 500 bases past the most distal 3' termini available in Ensembl v65, and collectively they add 6.6 Mb and 5.1 Mb to the mRNA space of mouse and human, respectively. Extensive Northern analyses validated stable accumulation of distal APA isoforms, including transcripts bearing exceptionally long 3' UTRs (many >10 kb and some >18 kb in length). The Northern data further illustrate that the extensions we annotated were not due to unprocessed transcriptional run-off events. Global tissue comparisons revealed that APA events yielding these extensions were most prevalent in the mouse and human brain. Finally, these extensions collectively contain thousands of conserved miRNA binding sites, and these are strongly enriched for many well-studied neural miRNAs. Altogether, these new 3' UTR annotations greatly expand the scope of post-transcriptional regulatory networks in mammals, and have particular impact on the central nervous system.
Figures


Similar articles
-
Global patterns of tissue-specific alternative polyadenylation in Drosophila.Cell Rep. 2012 Mar 29;1(3):277-89. doi: 10.1016/j.celrep.2012.01.001. Cell Rep. 2012. PMID: 22685694 Free PMC article.
-
Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs.Nature. 2011 Jan 6;469(7328):97-101. doi: 10.1038/nature09616. Epub 2010 Nov 17. Nature. 2011. PMID: 21085120 Free PMC article.
-
A compendium of conserved cleavage and polyadenylation events in mammalian genes.Genome Res. 2018 Oct;28(10):1427-1441. doi: 10.1101/gr.237826.118. Epub 2018 Aug 24. Genome Res. 2018. PMID: 30143597 Free PMC article.
-
Alternative polyadenylation in the nervous system: to what lengths will 3' UTR extensions take us?Bioessays. 2014 Aug;36(8):766-77. doi: 10.1002/bies.201300174. Epub 2014 Jun 5. Bioessays. 2014. PMID: 24903459 Free PMC article. Review.
-
Emerging Roles for 3' UTRs in Neurons.Int J Mol Sci. 2020 May 12;21(10):3413. doi: 10.3390/ijms21103413. Int J Mol Sci. 2020. PMID: 32408514 Free PMC article. Review.
Cited by
-
Animal-APAdb: a comprehensive animal alternative polyadenylation database.Nucleic Acids Res. 2021 Jan 8;49(D1):D47-D54. doi: 10.1093/nar/gkaa778. Nucleic Acids Res. 2021. PMID: 32986825 Free PMC article.
-
Transcriptome-Wide Analysis of Interplay between mRNA Stability, Translation and Small RNAs in Response to Neuronal Membrane Depolarization.Int J Mol Sci. 2020 Sep 25;21(19):7086. doi: 10.3390/ijms21197086. Int J Mol Sci. 2020. PMID: 32992958 Free PMC article.
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells.Genome Biol. 2024 Jan 23;25(1):31. doi: 10.1186/s13059-023-03146-x. Genome Biol. 2024. PMID: 38263082 Free PMC article. Review.
-
APAatlas: decoding alternative polyadenylation across human tissues.Nucleic Acids Res. 2020 Jan 8;48(D1):D34-D39. doi: 10.1093/nar/gkz876. Nucleic Acids Res. 2020. PMID: 31586392 Free PMC article.
-
Implications of Poly(A) Tail Processing in Repeat Expansion Diseases.Cells. 2022 Feb 15;11(4):677. doi: 10.3390/cells11040677. Cells. 2022. PMID: 35203324 Free PMC article. Review.
References
-
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A 2010. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci 13: 291–301 - PubMed
-
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL 2011. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev 6: 29. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
